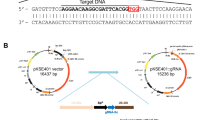
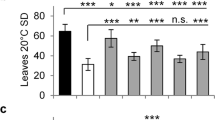
Abstract
The Arabidopsis genome encodes 29 AHL (AT-hook motif nuclear localized) proteins, but the function for most of them remains unknown. We report here a study of the AHL22 gene, which was originally identified as a gain-of-function allele that enhanced the phenotype of the cry1 cry2 mutant. AHL22 is a nuclear protein with the binding activity for an AT-rich DNA sequence. AHL22 overexpression delayed flowering and caused a constitutive photomorphogenic phenotype. The loss-of-function AHL22 mutant showed no clear phenotype on flowering, but slightly longer hypocotyls. However, silencing four AHL genes (AHL22, AHL18, AHL27, and AHL29) resulted in early flowering and enhanced ahl22-1 mutant phenotype on the growth of hypocotyls, suggesting genetic redundancy of AHL22 with other AHL genes on these plant developmental events. Further analysis showed that AHL22 controlled flowering and hypocotyl elongation might result from primarily the regulation of FT and PIF4 expression, respectively.






Similar content being viewed by others
References
Ahmad M, Cashmore AR (1993) HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366:162–166. doi:10.1038/366162a0
Aravind L, Landsman D (1998) AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res 26:4413–4421. doi:10.1093/nar/26.19.4413
Bastow R, Dean C (2003) Plant sciences. Deciding when to flower. Science 302:1695–1696. doi:10.1126/science.1092862
Cattaruzzi G, Altamura S, Tessari MA, Rustighi A, Giancotti V, Pucillo C, Manfioletti G (2007) The second AT-hook of the architectural transcription factor HMGA2 is determinant for nuclear localization and function. Nucleic Acids Res 35:1751–1760. doi:10.1093/nar/gkl1106
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743. doi:10.1046/j.1365-313x.1998.00343.x
Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR (2000) Random GFP:cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci USA 97:3718–3723. doi:10.1073/pnas.97.7.3718
El-Din El-Assal S, Alonso-Blanco C, Peeters AJ, Wagemaker C, Weller JL, Koornneef M (2003) The role of cryptochrome 2 in flowering in Arabidopsis. Plant Physiol 133:1504–1516. doi:10.1104/pp.103.029819
Ferrario S, Immink RG, Shchennikova A, Busscher-Lange J, Angenent GC (2003) The MADS box gene FBP2 is required for SEPALLATA function in petunia. Plant Cell 15:914–925. doi:10.1105/tpc.010280
Fujimoto S, Matsunaga S, Yonemura M, Uchiyama S, Azuma T, Fukui K (2004) Identification of a novel plant MAR DNA binding protein localized on chromosomal surfaces. Plant Mol Biol 56:225–239. doi:10.1007/s11103-004-3249-5
Grasser KD, Launholt D, Grasser M (2007a) High mobility group proteins of the plant HMGB family: dynamic chromatin modulators. Biochim Biophys Acta 1769:346–357
Grasser M, Christensen JM, Peterhansel C, Grasser KD (2007b) Basic and acidic regions flanking the HMG-box domain of maize HMGB1 and HMGB5 modulate the stimulatory effect on the DNA binding of transcription factor Dof2. Biochemistry 46:6375–6382. doi:10.1021/bi6024947
Guo H, Yang H, Mockler TC, Lin C (1998) Regulation of flowering time by Arabidopsis photoreceptors. Science 279:1360–1363. doi:10.1126/science.279.5355.1360
Harrer M, Luhrs H, Bustin M, Scheer U, Hock R (2004) Dynamic interaction of HMGA1a proteins with chromatin. J Cell Sci 117:3459–3471. doi:10.1242/jcs.01160
Huq E, Quail PH (2002) PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J 21:2441–2450. doi:10.1093/emboj/21.10.2441
Launholt D, Merkle T, Houben A, Schulz A, Grasser KD (2006) Arabidopsis chromatin-associated HMGA and HMGB use different nuclear targeting signals and display highly dynamic localization within the nucleus. Plant Cell 18:2904–2918. doi:10.1105/tpc.106.047274
LeClere S, Bartel B (2001) A library of Arabidopsis 35S-cDNA lines for identifying novel mutants. Plant Mol Biol 46:695–703. doi:10.1023/A:1011699722052
Lim PO, Kim Y, Breeze E, Koo JC, Woo HR, Ryu JS, Park DH, Beynon J, Tabrett A, Buchanan-Wollaston V, Nam HG (2007) Overexpression of a chromatin architecture-controlling AT-hook protein extends leaf longevity and increases the post-harvest storage life of plants. Plant J 52:1140–1153
Lin C, Ahmad M, Cashmore AR (1996) Arabidopsis cryptochrome 1 is a soluble protein mediating blue light-dependent regulation of plant growth and development. Plant J 10:893–902. doi:10.1046/j.1365-313X.1996.10050893.x
Lin C, Yang H, Guo H, Mockler T, Chen J, Cashmore AR (1998) Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc Natl Acad Sci USA 95:2686–2690. doi:10.1073/pnas.95.5.2686
Lu Q (2005) Seamless cloning and gene fusion. Trends Biotechnol 23:199–207. doi:10.1016/j.tibtech.2005.02.008
Martinez-Garcia JF, Quail PH (1999) The HMG-I/Y protein PF1 stimulates binding of the transcriptional activator GT-2 to the PHYA gene promoter. Plant J 18:173–183. doi:10.1046/j.1365-313X.1999.00440.x
Matsushita A, Furumoto T, Ishida S, Takahashi Y (2007) AGF1, an AT-hook protein, is necessary for the negative feedback of AtGA3ox1 encoding GA 3-oxidase. Plant Physiol 143:1152–1162. doi:10.1104/pp.106.093542
Mockler TC, Guo H, Yang H, Duong H, Lin C (1999) Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development 126:2073–2082
Nagy F, Schafer E (2002) Phytochromes control photomorphogenesis by differentially regulated, interacting signaling pathways in higher plants. Annu Rev Plant Biol 53:329–355. doi:10.1146/annurev.arplant.53.100301.135302
Ni M, Tepperman JM, Quail PH (1998) PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95:657–667. doi:10.1016/S0092-8674(00)81636-0
Ni M, Tepperman JM, Quail PH (1999) Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature 400:781–784. doi:10.1038/23500
Osterlund MT, Hardtke CS, Wei N, Deng XW (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405:462–466. doi:10.1038/35013076
Pelaz S, Gustafson-Brown C, Kohalmi SE, Crosby WL, Yanofsky MF (2001) APETALA1 and SEPALLATA3 interact to promote flower development. Plant J 26:385–394. doi:10.1046/j.1365-313X.2001.2641042.x
Saijo Y, Sullivan JA, Wang H, Yang J, Shen Y, Rubio V, Ma L, Hoecker U, Deng XW (2003) The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev 17:2642–2647. doi:10.1101/gad.1122903
Sgarra R, Lee J, Tessari MA, Altamura S, Spolaore B, Giancotti V, Bedford MT, Manfioletti G (2006) The AT-hook of the chromatin architectural transcription factor high mobility group A1a is arginine-methylated by protein arginine methyltransferase 6. J Biol Chem 281:3764–3772. doi:10.1074/jbc.M510231200
Street IH, Shah PK, Smith AM, Avery N, Neff MM (2008) The AT-hook-containing proteins SOB3/AHL29 and ESC/AHL27 are negative modulators of hypocotyl growth in Arabidopsis. Plant J 54:1–14. doi:10.1111/j.1365-313X.2007.03393.x
Su Y, Kwon CS, Bezhani S, Huvermann B, Chen C, Peragine A, Kennedy JF, Wagner D (2006) The N-terminal ATPase AT-hook-containing region of the Arabidopsis chromatin-remodeling protein SPLAYED is sufficient for biological activity. Plant J 46:685–699. doi:10.1111/j.1365-313X.2006.02734.x
Sung S, Amasino RM (2004) Vernalization and epigenetics: how plants remember winter. Curr Opin Plant Biol 7:4–10. doi:10.1016/j.pbi.2003.11.010
Turck F, Fornara F, Coupland G (2008) Regulation and identity of florigen: flowering locus T moves center stage. Annu Rev Plant Biol 59:573–594. doi:10.1146/annurev.arplant.59.032607.092755
Ulm R, Baumann A, Oravecz A, Mate Z, Adam E, Oakeley EJ, Schafer E, Nagy F (2004) Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc Natl Acad Sci USA 101:1397–1402. doi:10.1073/pnas.0308044100
Vom Endt D, Soares e Silva M, Kijne JW, Pasquali G, Memelink J (2007) Identification of a bipartite jasmonate-responsive promoter element in the Catharanthus roseus ORCA3 transcription factor gene that interacts specifically with AT-Hook DNA-binding proteins. Plant Physiol 144:1680–1689. doi:10.1104/pp.107.096115
Weigel D, Glazebrook J (2002) TAIL-PCR. In: Weigel D, Glazebrook J (eds) Arabidopsis, a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, pp 144–153
Weigel D, Ahn JH, Blazquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrandiz C, Kardailsky I, Malancharuvil EJ, Neff MM, Nguyen JT, Sato S, Wang ZY, Xia Y, Dixon RA, Harrison MJ, Lamb CJ, Yanofsky MF, Chory J (2000) Activation tagging in Arabidopsis. Plant Physiol 122:1003–1013. doi:10.1104/pp.122.4.1003
Zhao X, Yu X, Foo E, Symons GM, Lopez J, Bendehakkalu KT, Xiang J, Weller JL, Liu X, Reid JB, Lin C (2007) A study of gibberellin homeostasis and cryptochrome-mediated blue light inhibition of hypocotyl elongation. Plant Physiol 145:106–118. doi:10.1104/pp.107.099838
Acknowledgments
Thanks to Drs. Bekir Ülker and Jane Parker for kindly providing pJawohl8-RNAi and pLeela vectors, and to Dr. Robert Koebner for his critical reading of the manuscript. This work was supported in part by the National ‘‘863’’ Program of China (2006AA10A111, 2006AA10Z107, and 2007AA10Z119), the National Key Basic Research ‘973’ Program of China (2004CB117206), the Key Technology R & D Program (2007BAD59B02), National Science Foundation of China (30671245), National Institute of Health (GM56265), and UCLA faculty research and Sol Leshin BGU-UCLA Academic Cooperation programs.
Author information
Authors and Affiliations
Corresponding authors
Additional information
C. Xiao, F. Chen and X. Yu contributed equally to the work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xiao, C., Chen, F., Yu, X. et al. Over-expression of an AT-hook gene, AHL22, delays flowering and inhibits the elongation of the hypocotyl in Arabidopsis thaliana . Plant Mol Biol 71, 39–50 (2009). https://doi.org/10.1007/s11103-009-9507-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-009-9507-9