Abstract
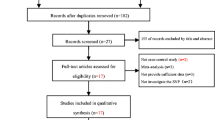
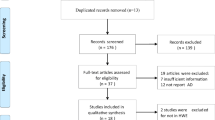
Whether the variations in the hemochromatosis (HFE) gene increase Alzheimer’s disease (AD) risk is still undetermined. We performed a meta-analysis in order to systematically summarize the possible association. Studies were identified by searching PUBMED, Web of Science and EMBASE databases complemented with screening the references of the retrieved studies. The association was measured using random-effect or fixed-effect odds ratio (OR) combined with 95% confidence intervals (CIs) according to the studies’ heterogeneity. For C282Y polymorphism, we did not find any association using data from 22 studies including 4,365 cases and 8,652 controls. For H63D polymorphism, on the basis of 2,795 cases and 7,424 controls from 17 studies, we observed a significant association (allele contrast: OR = 0.902, 95% CI = 0.819–0.994, P = 0.037; minor-allele-dominant model: OR = 0.887, 95% CI = 0.790–0.996, P = 0.043). No publication bias was detected in this meta-analysis. The synthesis of available evidence supports mutant of HFE H63D polymorphism plays a protective role for AD risk.



Similar content being viewed by others
References
Smith M, Harris P, Sayre L, Perry G (1997) Iron accumulation in Alzheimer disease is a source of redox-generated free radicals. Proc Natl Acad Sci USA 94:9866
Smith MA, Perry G (1995) Free radical damage, iron, and Alzheimer’s disease. J Neurol Sci 134(Suppl):92–94
Kala S, Hasinoff B, Richardson J (1996) Brain samples from Alzheimer’s patients have elevated levels of loosely bound iron. Int J Neurosci 86:263–269
Bothwell TH, MacPhail AP (1998) Hereditary hemochromatosis: etiologic, pathologic, and clinical aspects. Semin Hematol 35:55–71
Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22:719–748
DerSimonian R, Kacker R (2007) Random-effects model for meta-analysis of clinical trials: an update. Contemp Clin Trials 28:105–114. doi:10.1016/j.cct.2006.04.004
DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7:177–188
Kauwe J, Bertelsen S, Mayo K, Cruchaga C, Abraham R, Hollingworth P, Harold D, Owen M, Williams J, Lovestone S (2010) Suggestive synergy between genetic variants in TF and HFE as risk factors for Alzheimers disease. Am J Med Genet B Neuropsychiatr Genet 153B(4):955–959
Nandar W, Connor JR (2011) HFE gene variants affect iron in the brain. J Nutr 141:729S–739S. doi:10.3945/jn.110.130351
Ajioka RS, Levy JE, Andrews NC, Kushner JP (2002) Regulation of iron absorption in HFE mutant mice. Blood 100:1465–1469. doi:10.1182/blood-2001-11-0037
Parkkila S, Niemela O, Britton RS, Fleming RE, Waheed A, Bacon BR, Sly WS (2001) Molecular aspects of iron absorption and HFE expression. Gastroenterology 121:1489–1496
Chua AC, Drake SF, Herbison CE, Olynyk JK, Leedman PJ, Trinder D (2006) Limited iron export by hepatocytes contributes to hepatic iron-loading in the Hfe knockout mouse. J Hepatol 44:176–182. doi:10.1016/j.jhep.2005.08.012
Fleming RE, Britton RS (2006) Iron Imports VI. HFE and regulation of intestinal iron absorption. Am J Physiol Gastrointest Liver Physiol 290:G590–G594. doi:10.1152/ajpgi.00486.2005
Moalem S, Percy ME, Andrews DF, Kruck TP, Wong S, Dalton AJ, Mehta P, Fedor B, Warren AC (2000) Are hereditary hemochromatosis mutations involved in Alzheimer disease? Am J Med Genet 93:58–66. doi:10.1002/1096-8628(20000703)93:1<58:AID-AJMG10>3.0.CO;2-L
Chen C, Ma X, Zhong M, Yu Z (2010) Extremely low-frequency electromagnetic fields exposure and female breast cancer risk: a meta-analysis based on 24338 cases and 60628 controls. Breast Cancer Res Treat 123:569–576. doi:10.1007/s10549-010-0782-6
Ma X, Chen C, Xiong H, Fan J, Li Y, Lin H, Xu R, Huang G, Xu B (2010) No association between SOD2 Val16Ala polymorphism and breast cancer susceptibility: a meta-analysis based on 9,710 cases and 11,041 controls. Breast Cancer Res Treat 122:509–514. doi:10.1007/s10549-009-0725-2
Ma X, Chen C, Xiong H, Li Y (2010) Transforming growth factorbeta1 L10P variant plays an active role on the breast cancer susceptibility in Caucasian: evidence from 10,392 cases and 11,697 controls. Breast Cancer Res Treat 124:453–457. doi:10.1007/s10549-010-0843-x
Ma X, Qi X, Chen C, Lin H, Xiong H, Li Y, Jiang J (2010) Association between CYP19 polymorphisms and breast cancer risk: results from 10,592 cases and 11,720 controls. Breast Cancer Res Treat 122:495–501. doi:10.1007/s10549-009-0693-6
Sergentanis TN, Economopoulos KP (2010) Cyclin D1 G870A polymorphism and breast cancer risk: a meta-analysis comprising 9,911 cases and 11,171 controls. Mol Biol Rep. doi:10.1007/s11033-010-0639-4
Lehmann DJ, Schuur M, Warden DR, Hammond N, Belbin O, Kolsch H, Lehmann MG, Wilcock GK, Brown K, Kehoe PG, Morris CM, Barker R, Coto E, Alvarez V, Deloukas P, Mateo I, Gwilliam R, Combarros O, Arias-Vasquez A, Aulchenko YS, Ikram MA, Breteler MM, van Duijn CM, Oulhaj A, Heun R, Cortina-Borja M, Morgan K, Robson K, Smith AD (2010) Transferrin and HFE genes interact in Alzheimer’s disease risk: the Epistasis Project. Neurobiol Aging. doi:10.1016/j.neurobiolaging.2010.07.018
Correia AP, Pinto JP, Dias V, Mascarenhas C, Almeida S, Porto G (2009) CAT53 and HFE alleles in Alzheimer’s disease: a putative protective role of the C282Y HFE mutation. Neurosci Lett 457:129–132. doi:10.1016/j.neulet.2009.03.088
Li H, Wetten S, Li L, St Jean P, Upmanyu R, Surh L, Hosford D, Barnes M, Briley J, Borrie M (2008) Candidate single-nucleotide polymorphisms from a genomewide association study of Alzheimer disease. Arch Neurol 65:45
Percy M, Moalem S, Garcia A, Somerville MJ, Hicks M, Andrews D, Azad A, Schwarz P, Beheshti Zavareh R, Birkan R, Choo C, Chow V, Dhaliwal S, Duda V, Kupferschmidt AL, Lam K, Lightman D, Machalek K, Mar W, Nguyen F, Rytwinski PJ, Svara E, Tran M, Wheeler K, Yeung L, Zanibbi K, Zener R, Ziraldo M, Freedman M (2008) Involvement of ApoE E4 and H63D in sporadic Alzheimer’s disease in a folate-supplemented Ontario population. J Alzheimers Dis 14:69–84
Reiman EM, Webster JA, Myers AJ, Hardy J, Dunckley T, Zismann VL, Joshipura KD, Pearson JV, Hu-Lince D, Huentelman MJ, Craig DW, Coon KD, Liang WS, Herbert RH, Beach T, Rohrer KC, Zhao AS, Leung D, Bryden L, Marlowe L, Kaleem M, Mastroeni D, Grover A, Heward CB, Ravid R, Rogers J, Hutton ML, Melquist S, Petersen RC, Alexander GE, Caselli RJ, Kukull W, Papassotiropoulos A, Stephan DA (2007) GAB2 alleles modify Alzheimer’s risk in APOE epsilon4 carriers. Neuron 54:713–720. doi:10.1016/j.neuron.2007.05.022
Blazquez L, De Juan D, Ruiz-Martinez J, Emparanza JI, Saenz A, Otaegui D, Sistiaga A, Martinez-Lage P, Lamet I, Samaranch L, Buiza C, Etxeberria I, Arriola E, Cuadrado E, Urdaneta E, Yanguas J, Lopez de Munain A (2007) Genes related to iron metabolism and susceptibility to Alzheimer’s disease in Basque population. Neurobiol Aging 28:1941–1943. doi:10.1016/j.neurobiolaging.2006.08.009
Guerreiro RJ, Bras JM, Santana I, Januario C, Santiago B, Morgadinho AS, Ribeiro MH, Hardy J, Singleton A, Oliveira C (2006) Association of HFE common mutations with Parkinson’s disease, Alzheimer’s disease and mild cognitive impairment in a Portuguese cohort. BMC Neurol 6:24. doi:10.1186/1471-2377-6-24
Berlin D, Chong G, Chertkow H, Bergman H, Phillips N, Schipper H (2004) Evaluation of HFE (hemochromatosis) mutations as genetic modifiers in sporadic AD and MCI. Neurobiol Aging 25:465–474
Candore G, Licastro F, Chiappelli M, Franceschi C, Lio D, Balistreri CR, Piazza G, Colonna-Romano G, Grimaldi LM, Caruso C (2003) Association between the HFE mutations and unsuccessful ageing: a study in Alzheimer’s disease patients from Northern Italy. Mech Ageing Dev 124:525–528. doi:10.1016/S0047-6374(03)00031-9
Pulliam JF, Jennings CD, Kryscio RJ, Davis DG, Wilson D, Montine TJ, Schmitt FA, Markesbery WR (2003) Association of HFE mutations with neurodegeneration and oxidative stress in Alzheimer’s disease and correlation with APOE. Am J Med Genet B Neuropsychiatr Genet 119B:48–53. doi:10.1002/ajmg.b.10069
Lleo A, Blesa R, Angelopoulos C, Pastor-Rubio P, Villa M, Oliva R, Bufill E (2002) Transferrin C2 allele, haemochromatosis gene mutations, and risk for Alzheimer’s disease. J Neurol Neurosurg Psychiatry 72:820–821
Sampietro M, Caputo L, Casatta A, Meregalli M, Pellagatti A, Tagliabue J, Annoni G, Vergani C (2001) The hemochromatosis gene affects the age of onset of sporadic Alzheimer’s disease. Neurobiol Aging 22:563–568
Acknowledgments
We thank Dr Kauwe from Department of Biology, Brigham Young University, for providing us the detailed information about their study; Dr Lehmann from Oxford Project to Investigate Memory and Ageing (OPTIMA), University Department of Physiology, Anatomy and Genetics, Oxford, UK, and Dr Ibrahim from Department of Epidemiology, Erasmus MC University Medical Center, Rotterdam, the Netherlands, for interpreting the overlapping dada via e-mail during the manuscript writing.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Min Lin and Lin Zhao contributed equally to this article.
Rights and permissions
About this article
Cite this article
Lin, M., Zhao, L., Fan, J. et al. Association between HFE polymorphisms and susceptibility to Alzheimer’s disease: a meta-analysis of 22 studies including 4,365 cases and 8,652 controls. Mol Biol Rep 39, 3089–3095 (2012). https://doi.org/10.1007/s11033-011-1072-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-011-1072-z