Abstract
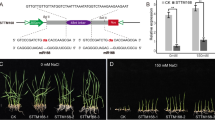
Salinity and alkalinity are the two main environmental factors that limit rice production. Better understanding of the mechanisms responsible for salinity and alkaline stress tolerance would allow researchers to modify rice to increase its resistance to salinity and alkaline stress. MicroRNAs (miRNAs) are ~21-nucleotide RNAs that are ubiquitous regulators of gene expression in eukaryotic organisms. Some miRNAs acts as an important endogenous regulator in plant responses to abiotic stressors. miR393 is a conservative miRNA family that occurs in a variety of different plants. The two members of the miR393 family found in rice are named osa-MIR393 and osa-MIR393b. We found that the osa-MIR393 expression level changed under salinity and alkaline stress, whereas that of osa-MIR393b did not. Target genes of osa-MIR393 were predicted, and some of these putative targets are abiotic related genes. Furthermore, we generated transgenic rice and Arabidopsis thaliana that over-expressed osa-MIR393, and the phenotype analysis showed that these transgenic plants were more sensitive to salt and alkali treatment compared to wild-type plants. These results illustrate that over-expression of osa-MIR393 can negatively regulate rice salt-alkali stress tolerance.




Similar content being viewed by others
Abbreviations
- miRNA:
-
MicroRNA
- GRF:
-
Growth-regulating factor
- GIF:
-
GRF-interacting factor
- MS medium:
-
Murashige and Skoog medium
- PCR:
-
Polymerase chain reaction
- RT-PCR:
-
Reverse transcription PCR
- pre-miRNA:
-
miRNA precursor
- GFP:
-
Green fluorescent protein
- RACE:
-
Rapid-amplification of cDNA ends
References
Bartel B, Bartel DP (2003) MicroRNAs: at the root of plant development? Plant Physiol 132:709–717
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297
Lai EC (2003) MicroRNAs: runts of the genome assert themselves. Curr Biol 13:R925–R936
Mallory AC, Vaucheret H (2006) Functions of microRNAs and related small RNAs in plants. Nat Genet Suppl 38:S31–S36
Sunkar R, Zhu JK (2004) Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16(8):2001–2019
Jones-Rhoades MW, Bartel DP (2004) Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14(6):787–799
Zhao B, Liang R, Ge L, Li W, Xiao H, Lin H, Ruan K, Jin Y (2007) Identification of drought-induced microRNAs in rice. Biochem Biophys Res Commun 354(2):585–590
Yoshida S, Forno DA, Cock JH, Gomez KA (1976) Laboratory manual for physiological studies of rice. IRRI, Los Banos
Chakrabarty R, Banerjee R, Chung SM, Farman M, Citovsky V, Hogenhout SA, Tzfira T, Goodin M (2007) pSITE vectors for stable integration or transient expression of autofluorescent protein fusions in plants: probing Nicotiana benthamiana-virus interactions. Mol Plant Microbe Interact 20:740–750
Voinnet O, Baulcombe DC (1997) Systemic signalling in gene silencing. Nature 389:553
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Toki S (1997) Rapid and efficient Agrobacterium-mediated transformation in rice. Plant Mol Biol Rep 15:16–21
Huang J, Zhang H, Wang J, Yang J (2003) Molecular cloning and characterization of rice 6-phosphogluconate dehydrogenase gene that is up-regulated by salt stress. Mol Biol Rep 30(4):223–227
Yang W, Liu X, Zhang J, Feng J, Li C, Chen J (2009) Prediction and validation of conservative microRNAs of Solanum tuberosum L. Mol Biol Rep. doi:10.1007/s11033-009-9881-z
Wang YC, Qu GZ, Li HY, Wu YJ, Wang C, Liu GF, Yang CP (2010) Enhanced salt tolerance of transgenic poplar plants expressing a manganese superoxide dismutase from Tamarix androssowii. Mol Biol Rep 37(2):1119–1124
Sunkar R, Chinnusamy V, Zhu J, Zhu JK (2007) Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci 12:301–309
Phillips JR, Dalmay T, Bartels D (2007) The role of small RNAs in abiotic stress. FEBS Lett 581:3592–3597
Sunkar R, Jagadeeswaran G (2008) In silico identification of conserved microRNAs in large number of diverse plant species. BMC Plant Biol 8:37
Liu HH, Tian X, Li YJ et al (2008) Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 14:836–843
Liu C, Zhang L, Sun J, Luo Y, Wang MB, Fan YL, Wang L (2010) A simple artificial microRNA vector based on ath-miR169d precursor from Arabidopsis. Mol Biol Rep 37:903–909
Acknowledgments
This project was supported by a grant from the Key Research Plan of Heilongjiang Province (GA06B103-3), the Innovation Research Group of NEAU (CXT004), the “863” project (2008AA10Z153), and the Basic Research Preliminary Study Foundation of the Ministry of Science and Technology of the PRC (2003CCA03500).
Author information
Authors and Affiliations
Corresponding author
Additional information
Peng Gao and Xi Bai contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gao, P., Bai, X., Yang, L. et al. osa-MIR393: a salinity- and alkaline stress-related microRNA gene. Mol Biol Rep 38, 237–242 (2011). https://doi.org/10.1007/s11033-010-0100-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-010-0100-8