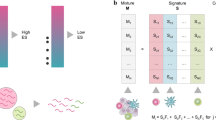
Abstract
Tumor infiltrating leukocytes (TILs) are an integral component of the tumor microenvironment and have been found to correlate with prognosis and response to therapy. Methods to enumerate immune subsets such as immunohistochemistry or flow cytometry suffer from limitations in phenotypic markers and can be challenging to practically implement and standardize. An alternative approach is to acquire aggregative high dimensional data from cellular mixtures and to subsequently infer the cellular components computationally. We recently described CIBERSORT, a versatile computational method for quantifying cell fractions from bulk tissue gene expression profiles (GEPs). Combining support vector regression with prior knowledge of expression profiles from purified leukocyte subsets, CIBERSORT can accurately estimate the immune composition of a tumor biopsy. In this chapter, we provide a primer on the CIBERSORT method and illustrate its use for characterizing TILs in tumor samples profiled by microarray or RNA-Seq.
Similar content being viewed by others
References
Fridman WH, Pagès F, Sautès-Fridman C, Galon J (2012) The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer 12(4):298–306. https://doi.org/10.1038/nrc3245
Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, Nair VS, Xu Y, Khuong A, Hoang CD, Diehn M, West RB, Plevritis SK, Alizadeh AA (2015) The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med 21(8):938–945. https://doi.org/10.1038/nm.3909
Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu V, West AN, Carmona M, Kivork C, Seja E, Cherry G, Gutierrez AJ, Grogan TR, Mateus C, Tomasic G, Glaspy JA, Emerson RO, Robins H, Pierce RH, Elashoff DA, Robert C, Ribas A (2014) PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515(7528):568–571. https://doi.org/10.1038/nature13954
Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS, Sosman JA, McDermott DF, Powderly JD, Gettinger SN, Kohrt HE, Horn L, Lawrence DP, Rost S, Leabman M, Xiao Y, Mokatrin A, Koeppen H, Hegde PS, Mellman I, Chen DS, Hodi FS (2014) Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 515(7528):563–567. https://doi.org/10.1038/nature14011
Ji RR, Chasalow SD, Wang L, Hamid O, Schmidt H, Cogswell J, Alaparthy S, Berman D, Jure-Kunkel M, Siemers NO, Jackson JR, Shahabi V (2012) An immune-active tumor microenvironment favors clinical response to ipilimumab. Cancer Immunol Immunother 61(7):1019–1031. https://doi.org/10.1007/s00262-011-1172-6
Tung JW, Heydari K, Tirouvanziam R, Parks DR, Herzenberg LA, Herzenberg LA (2007) Modern flow cytometry: a practical approach. Clin Lab Med 27(3):453. https://doi.org/10.1016/j.cII.2007.05.001
Abbas AR, Wolslegel K, Seshasayee D, Modrusan Z, Clark HF (2009) Deconvolution of blood microarray data identifies cellular activation patterns in systemic lupus erythematosus. PLoS One 4(7):e6098. https://doi.org/10.1371/journal.pone.0006098
Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, Gräf S, Ha G, Haffari G, Bashashati A, Russell R, McKinney S, Langerød A, Green A, Provenzano E, Wishart G, Pinder S, Watson P, Markowetz F, Murphy L, Ellis I, Purushotham A, Børresen-Dale AL, Brenton JD, Tavaré S, Caldas C, Aparicio S, Group M (2012) The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486(7403):346–352. https://doi.org/10.1038/nature10983
Ascierto ML, Kmieciak M, Idowu MO, Manjili R, Zhao Y, Grimes M, Dumur C, Wang E, Ramakrishnan V, Wang XY, Bear HD, Marincola FM, Manjili MH (2012) A signature of immune function genes associated with recurrence-free survival in breast cancer patients. Breast Cancer Res Treat 131(3):871–880. https://doi.org/10.1007/s10549-011-1470-x
Mann GJ, Pupo GM, Campain AE, Carter CD, Schramm SJ, Pianova S, Gerega SK, De Silva C, Lai K, Wilmott JS, Synnott M, Hersey P, Kefford RF, Thompson JF, Yang YH, Scolyer RA (2013) BRAF mutation, NRAS mutation, and the absence of an immune-related expressed gene profile predict poor outcome in patients with stage III melanoma. J Invest Dermatol 133(2):509–517. https://doi.org/10.1038/jid.2012.283
Galon J, Angell HK, Bedognetti D, Marincola FM (2013) The continuum of cancer immunosurveillance: prognostic, predictive, and mechanistic signatures. Immunity 39(1):11–26. https://doi.org/10.1016/j.immuni.2013.07.008
Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pagès C, Tosolini M, Camus M, Berger A, Wind P, Zinzindohoué F, Bruneval P, Cugnenc PH, Trajanoski Z, Fridman WH, Pagès F (2006) Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 313(5795):1960–1964. https://doi.org/10.1126/science.1129139
Tosolini M, Kirilovsky A, Mlecnik B, Fredriksen T, Mauger S, Bindea G, Berger A, Bruneval P, Fridman WH, Pagès F, Galon J (2011) Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, th2, treg, th17) in patients with colorectal cancer. Cancer Res 71(4):1263–1271. https://doi.org/10.1158/0008-5472.can-10-2907
Verhaak RG, Tamayo P, Yang JY, Hubbard D, Zhang H, Creighton CJ, Fereday S, Lawrence M, Carter SL, Mermel CH, Kostic AD, Etemadmoghadam D, Saksena G, Cibulskis K, Duraisamy S, Levanon K, Sougnez C, Tsherniak A, Gomez S, Onofrio R, Gabriel S, Chin L, Zhang N, Spellman PT, Zhang Y, Akbani R, Hoadley KA, Kahn A, Köbel M, Huntsman D, Soslow RA, Defazio A, Birrer MJ, Gray JW, Weinstein JN, Bowtell DD, Drapkin R, Mesirov JP, Getz G, Levine DA, Meyerson M, Network CGAR (2013) Prognostically relevant gene signatures of high-grade serous ovarian carcinoma. J Clin Invest 123(1):517–525. https://doi.org/10.1172/jci65833
Liebner DA, Huang K, Parvin JD (2014) MMAD: microarray microdissection with analysis of differences is a computational tool for deconvoluting cell type-specific contributions from tissue samples. Bioinformatics 30(5):682–689. https://doi.org/10.1093/bioinformatics/btt566
Zhong Y, Wan YW, Pang K, Chow LM, Liu Z (2013) Digital sorting of complex tissues for cell type-specific gene expression profiles. BMC Bioinformatics 14:89. https://doi.org/10.1186/1471-2105-14-89
Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA (2015) Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 12(5):453–457. https://doi.org/10.1038/nmeth.3337
Newman AM, Alizadeh AA (2016) High-throughput genomic profiling of tumor-infiltrating leukocytes. Curr Opin Immunol 41:77–84. https://doi.org/10.1016/j.coi.2016.06.006
Scholkopf B, Smola AJ, Williamson RC, Bartlett PL (2000) New support vector algorithms. Neural Comput 12(5):1207–1245
Corces MR, Buenrostro JD, Wu BJ, Greenside PG, Chan SM, Koenig JL, Snyder MP, Pritchard JK, Kundaje A, Gkeenleaf WJ, Majeti R, Chang HY (2016) Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet 48(10):1193–1203. https://doi.org/10.1038/ng.3646
Linsley PS, Speake C, Whalen E, Chaussabel D (2014) Copy number loss of the interferon gene cluster in melanomas is linked to reduced T cell infiltrate and poor patient prognosis. PLoS One 9(10):Artn E109760. https://doi.org/10.1371/Journal.Pone.0109760
Sleasman JW, Leon BH, Aleixo LF, Rojas M, Goodenow MM (1997) Immunomagnetic selection of purified monocyte and lymphocyte populations from peripheral blood mononuclear cells following cryopreservation. Clin Diagn Lab Immunol 4(6):653–658
Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, Trevino V, Shen H, Laird PW, Levine DA, Carter SL, Getz G, Stemke-Hale K, Mills GB, Verhaak RGW (2013) Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 4:Artn 2612. https://doi.org/10.1038/Ncomms3612
Acknowledgments
We would like to thank David Steiner, M.D., Ph.D. for his assistance in generating the RNA-Seq derived signature matrix. This work is supported by grants from the Doris Duke Charitable Foundation (A.A.A.), the Damon Runyon Cancer Research Foundation (A.A.A.), the B&J Cardan Oncology Research Fund (A.A.A.), the Ludwig Institute for Cancer Research (A.A.A.), NIH grant 1K99CA187192-01A1 (A.M.N.), NIH grant PHS NRSA 5T32 CA09302-35 (A.M.N.), US Department of Defense grant W81XWH-12-1-0498 (A.M.N.), a grant from the Siebel Stem Cell Institute and the Thomas and Stacey Siebel Foundation (A.M.N.), an NIH/Stanford MSTP training grant (B.C.), and a PD Soros Fellowship (B.C.).
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Science+Business Media LLC
About this protocol
Cite this protocol
Chen, B., Khodadoust, M.S., Liu, C.L., Newman, A.M., Alizadeh, A.A. (2018). Profiling Tumor Infiltrating Immune Cells with CIBERSORT. In: von Stechow, L. (eds) Cancer Systems Biology. Methods in Molecular Biology, vol 1711. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-7493-1_12
Download citation
DOI: https://doi.org/10.1007/978-1-4939-7493-1_12
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-7492-4
Online ISBN: 978-1-4939-7493-1
eBook Packages: Springer Protocols