Note: c302 has recently been moved to its own repository!!
Old README:
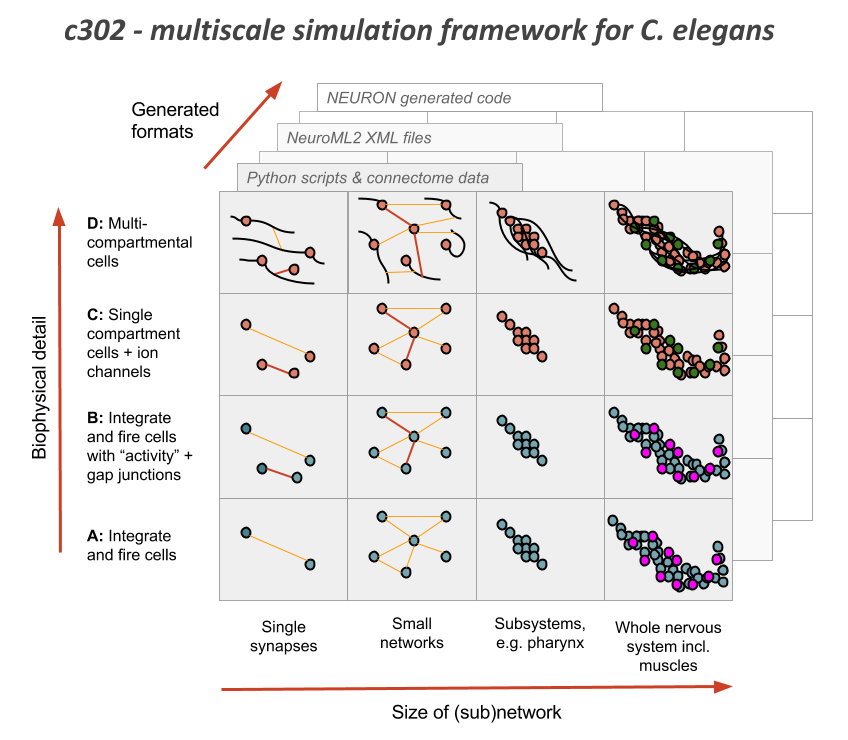
c302 is an experimental framework for generating network models in NeuroML 2 based on C elegans connectivity data.
It uses information on the synaptic connectivity of the network (from here) and uses libNeuroML to generate a network in valid NeuroML, which can be run in jNeuroML or pyNeuroML.
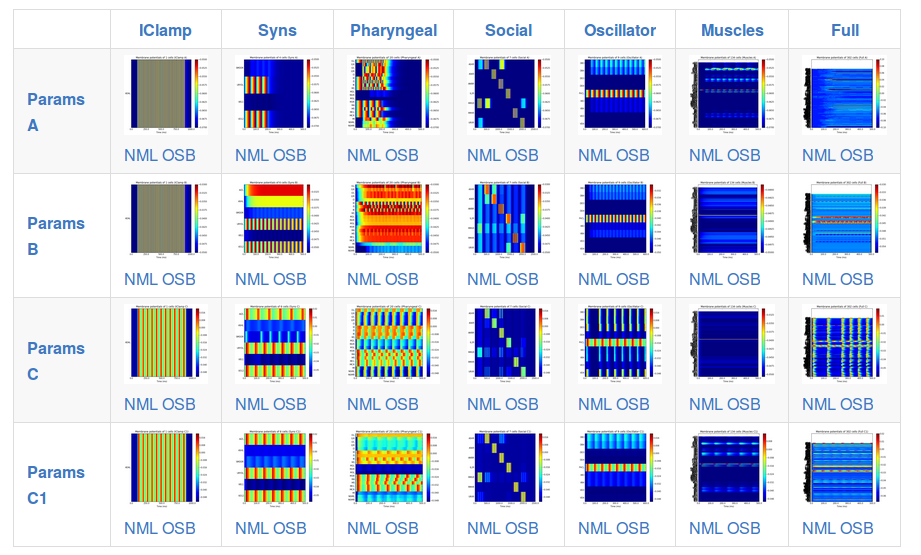
There will be multiple version of this network (see figure above), based on increasingly complex cell models, e.g.
Parameters_A Integrate & fire cells (not very physiological) connected by chemical (event triggered, conductance based) synapses
Parameters_B Updated I&F cells, with gap junction connections plus an "activity" measure (varies 0->1 depending on depolarisation of cell), which should be a better approximation for the relative activity of cells
Parameters_C Single compartment, conductance based neurons (will be initially based on muscle cell model). A modified version of this (Parameters_C1) has analogue (graded) synapses.
Parameters_D (TODO) Multicompartmental, conductance based cells
Parameters A, B and C/C1 are the only parameter sets tested so far, but the aim is to make all of the associated tools below for running, visualising, analysing, etc. independent of the parameter set used, so they can be ready for more detailed networks from c302 in the future.
The full set of dependencies for c302 can be installed with the following (see also the Travis-CI script):
python setup.py install
To regenerate a set of NeuroML & LEMS files for one instance of the model and execute it:
cd ./CElegans/pythonScripts/c302 # Enter c302 script directory
python c302_Full.py # To regenerate the NeuroML & LEMS files
pynml examples/LEMS_c302_A_Full.xml # Run a simulation with jNeuroML via pyNeuroML
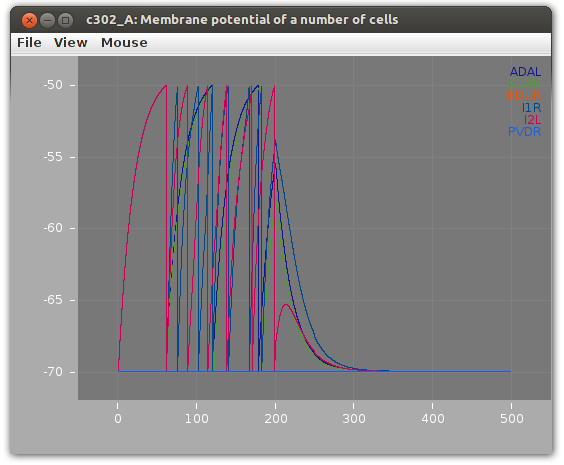
This will produce the following (6 cells visualised with the jNeuroML GUI):
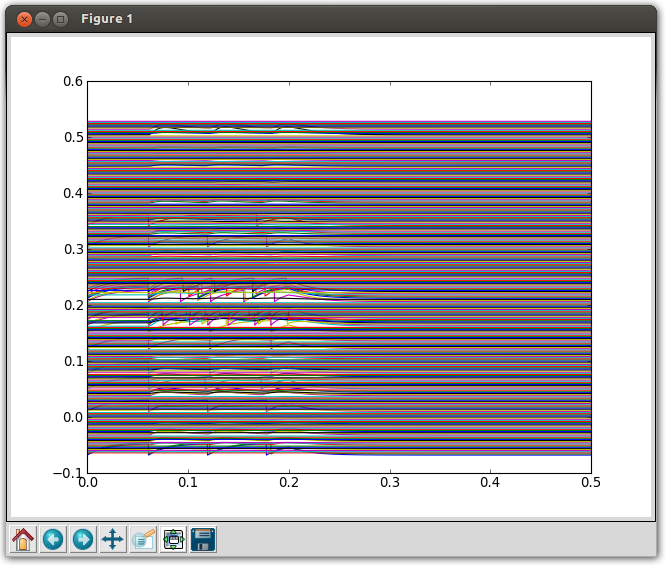
This saves traces from all neurons in a file c302_A.dat. To plot the membrane potential of all 302 neurons:
python analyse.py c302_A_Full.dat
Traces are slightly offset from one another for clarity.
To test all of the working features of the framework run test.sh:
./test.sh
This package can be used to generate customised networks of varying size, with different cells stimulated, of varying duration from the command line:
./c302.py MyNetwork parameters_A -cells ["ADAL","AIBL","RIVR","RMEV"] -cellstostimulate ["ADAL","RIVR"] -duration 500
This will create a NeuroML 2 file and a LEMS file to execute it, containing 4 cells, stimulating 2 of them, and with a duration of 500 ms
More options can be found with
./c302.py -h
Due to the fact that the cells are in pure NeuroML2, they can be mapped to other formats using the export feature of jNeuroML. Install NEURON and map the network to this format using:
cd examples
for jNeuroML:
jnml LEMS_c302_A_Pharyngeal.xml -neuron
or instead for pyNeuroML:
pynml LEMS_c302_A_Pharyngeal.xml -neuron
then
nrnivmodl
nrngui -python LEMS_c302_A_Pharyngeal_nrn.py
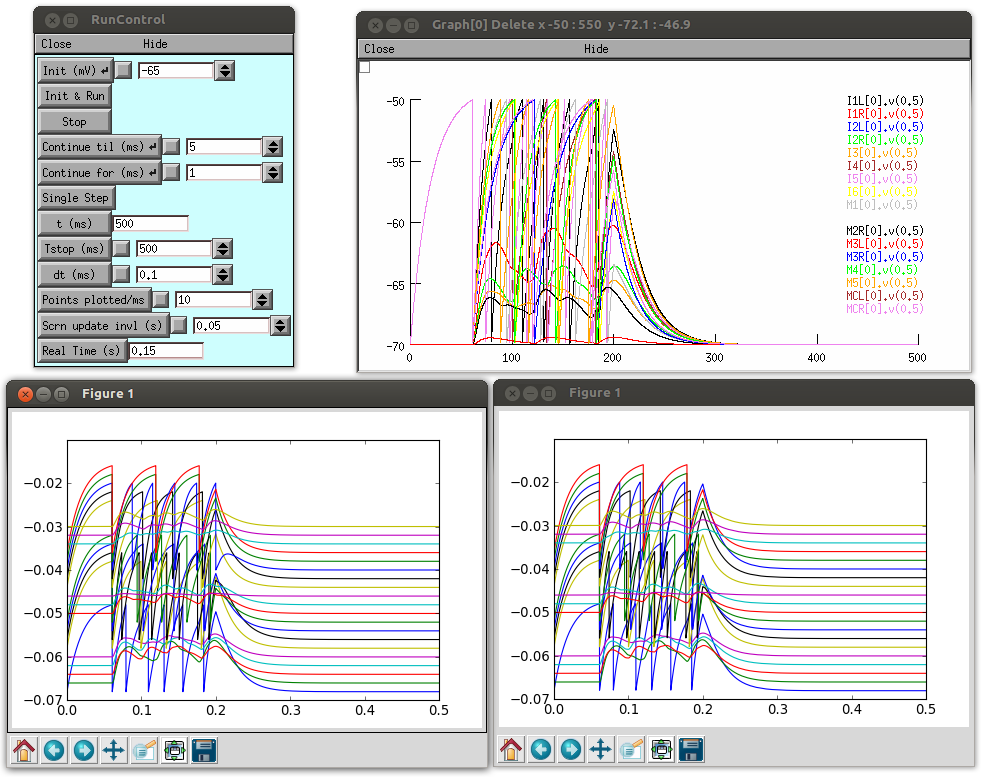
This will run the example network containing just the 20 cells from the pharynx.
Image above shows the network run in NEURON (top) and a comparison of the activity of the 20 cells when run on jNeuroML (bottom left) and NEURON (bottom right). Bottom graphs generated with analyse.py and each of the traces are offset by a few mV for clarity.
See here for more details on this.
Future plans include:
-
Implement & correctly tune Parameters_C (started), Parameters_D
-
Modify to use PyOpenWorm as source of connection data
-
Move all of this to its own repository
-
Add to Geppetto for simulation of networks in browser(done, see live.geppetto.org) -
Add muscle cells & connect output to muscle cell activity in Sibernetic
-
Link to bionet to tune weights of network to physiological behaviour