Learning protein structural similarity from sequence alone. Our preprint can be found here
DeepBLAST can be installed from pip via
pip install deepblast
To install from the development branch run
pip install git+https://github.com/flatironinstitute/deepblast.git
The pretrained DeepBLAST model can be downloaded here.
The TM-align structural alignments used to pretrain DeepBLAST can be found here
See the Malisam and Malidup websites to download their datasets.
We have 2 command line scripts available, namely deepblast-train and deepblast-eval.
deepblast-train takes in as input a tab-delimited format of with columns
query_seq_id | key_seq_id | tm_score1 | tm_score2 | rmsd | sequence1 | sequence2 | alignment_string
See an example here of what this looks like. At this moment, we only support parsing the output of TM-align. The parsing script can be found under
deepblast/dataset/parse_tm_align.py [fname] [output_table]
Once the data is configured and split appropriately, deepblast-train can be run.
The command-line options are given below (see deepblast-train --help for more details).
usage: deepblast-train [-h] [--gpus GPUS] [--grad-accum GRAD_ACCUM] [--grad-clip GRAD_CLIP] [--nodes NODES] [--num-workers NUM_WORKERS] [--precision PRECISION] [--backend BACKEND]
[--load-from-checkpoint LOAD_FROM_CHECKPOINT] --train-pairs TRAIN_PAIRS --test-pairs TEST_PAIRS --valid-pairs VALID_PAIRS [--embedding-dim EMBEDDING_DIM]
[--rnn-input-dim RNN_INPUT_DIM] [--rnn-dim RNN_DIM] [--layers LAYERS] [--loss LOSS] [--learning-rate LEARNING_RATE] [--batch-size BATCH_SIZE]
[--multitask MULTITASK] [--finetune FINETUNE] [--mask-gaps MASK_GAPS] [--scheduler SCHEDULER] [--epochs EPOCHS]
[--visualization-fraction VISUALIZATION_FRACTION] -o OUTPUT_DIRECTORY
This will evaluate how much the deepblast predictions agree with the structural alignments.
The deepblast-train command will automatically evaluate the heldout test set if it completes.
However, a separate deepblast-evaluate command is available in case the pretraining was interrupted. The commandline options are given below (see deepblast-evaluate --help for more details)
usage: deepblast-evaluate [-h] [--gpus GPUS] [--num-workers NUM_WORKERS] [--nodes NODES] [--load-from-checkpoint LOAD_FROM_CHECKPOINT] [--precision PRECISION] [--backend BACKEND]
--train-pairs TRAIN_PAIRS --test-pairs TEST_PAIRS --valid-pairs VALID_PAIRS [--embedding-dim EMBEDDING_DIM] [--rnn-input-dim RNN_INPUT_DIM]
[--rnn-dim RNN_DIM] [--layers LAYERS] [--loss LOSS] [--learning-rate LEARNING_RATE] [--batch-size BATCH_SIZE] [--multitask MULTITASK]
[--finetune FINETUNE] [--mask-gaps MASK_GAPS] [--scheduler SCHEDULER] [--epochs EPOCHS] [--visualization-fraction VISUALIZATION_FRACTION] -o
OUTPUT_DIRECTORY
import torch
from deepblast.trainer import LightningAligner
from deepblast.dataset.utils import pack_sequences
from deepblast.dataset.utils import states2alignment
import matplotlib.pyplot as plt
import seaborn as sns
# Load the pretrained model
model = LightningAligner.load_from_checkpoint(your_model_path)
# Load on GPU (if you want)
model = model.cuda()
# Obtain hard alignment from the raw sequences
x = 'IGKEEIQQRLAQFVDHWKELKQLAAARGQRLEESLEYQQFVANVEEEEAWINEKMTLVASED'
y = 'QQNKELNFKLREKQNEIFELKKIAETLRSKLEKYVDITKKLEDQNLNLQIKISDLEKKLSDA'
pred_alignment = model.align(x, y)
x_aligned, y_aligned = states2alignment(pred_alignment, x, y)
print(x_aligned)
print(pred_alignment)
print(y_aligned)
x_ = torch.Tensor(model.tokenizer(str.encode(x))).long()
y_ = torch.Tensor(model.tokenizer(str.encode(y))).long()
# Pack sequences for easier parallelization
seq, order = pack_sequences([x_], [y_])
seq = seq.cuda()
# Generate alignment score
score = model.aligner.score(seq, order).item()
print('Score', score)
# Predict expected alignment
A, match_scores, gap_scores = model.forward(seq, order)
# Display the expected alignment
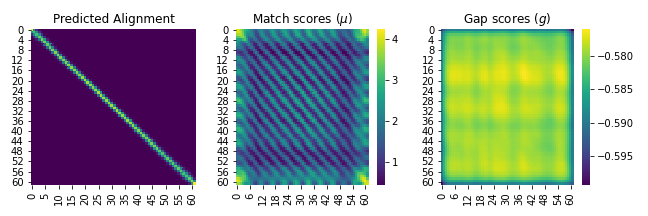
fig, ax = plt.subplots(1, 3, figsize=(9, 3))
sns.heatmap(A.cpu().detach().numpy().squeeze(), ax=ax[0], cbar=False, cmap='viridis')
sns.heatmap(match_scores.cpu().detach().numpy().squeeze(), ax=ax[1], cmap='viridis')
sns.heatmap(gap_scores.cpu().detach().numpy().squeeze(), ax=ax[2], cmap='viridis')
ax[0].set_title('Predicted Alignment')
ax[1].set_title('Match scores ($\mu$)')
ax[2].set_title('Gap scores ($g$)')
plt.tight_layout()
plt.show()The output will look like
IGKEEIQQRLAQFVDHWKELKQLAAARGQRLEESLEYQQFVANVEEEEAWINEKMTLVASED
::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::
QQNKELNFKLREKQNEIFELKKIAETLRSKLEKYVDITKKLEDQNLNLQIKISDLEKKLSDA
Score 282.3163757324219
Q : How do I interpret the alignment string?
A : The alignment string is used to indicate matches and mismatches between sequences. For example consider the following alignment
ADQSFLWASGVI-S------D-EM--
::::::::::::2:222222:2:122
MHHHHHHSSGVDLWSHPQFEKGT-EN
The first 12 residues in the alignment are matches. The last 2 characters indicate insertions in the second sequence (hence the 2 in the alignment string), and the 3rd to last character indciates an insertion in the first sequence (hence the 1 in the aligment string).
If you find our work useful, please cite us at
@article{morton2020protein,
title={Protein Structural Alignments From Sequence},
author={Morton, Jamie and Strauss, Charlie and Blackwell, Robert and Berenberg, Daniel and Gligorijevic, Vladimir and Bonneau, Richard},
journal={bioRxiv},
year={2020},
publisher={Cold Spring Harbor Laboratory}
}