This is an overview of all github repositories associated with PuckerLab. Please find a list of the science communication projects, the developed bioinformatics tools, teaching activities, and project specific repositories below.
Specific facts and research projects about MYB transcription factors are shared on a weekly basis via Twitter and LinkedIn using #MybMonday. All posts are collected in the #MybMonday github repository.
Specific facts and research projects about the flavonoid biosynthesis are shared on a weekly basis via Twitter and LinkedIn using #FlavonoidFriday. All posts are collected in the #FlavonoidFriday github repository.
Knowledge-based Identification of Pathway Enzymes (KIPEs) performs an automatic annotation of the flavonoid biosynthesis steps in a new transcriptome of genome sequence assembly. Scripts and datasets are availabe in the KIPEs repository. KIPEs is also available on our web server. This enables all scientists to use KIPEs in their research projects. Please find additional details in the corresponding publication 'Automatic Identification of Players in the Flavonoid Biosynthesis with Application on the Biomedicinal Plant Croton tiglium'. Recently, we updated KIPEs with additional features and data sets as described in our preprint 'KIPEs3: Automatic annotation of biosynthesis pathways'.
This tool performs an automatic identification, annotation, and analysis of the MYB gene family in plants. It can be applied to new transcriptome of genome assemblies. Please find the scripts and data sets in the MYB_annotator repository. The MYB_annotator is also available on our web server. This enables all scientists to use KIPEs in their research projects. Please find additional details in the corresponding publication 'Automatic identification and annotation of MYB gene family members in plants'.
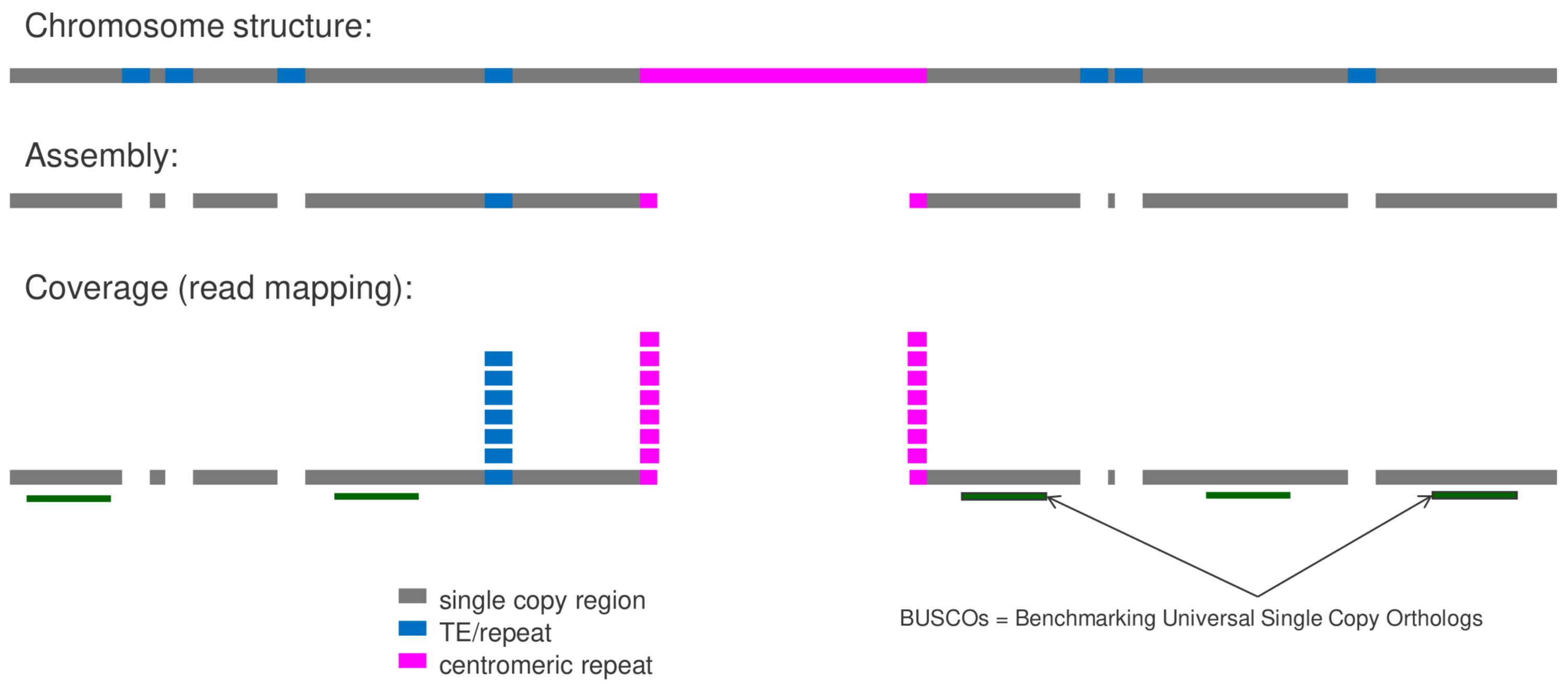
Mapping-based Genome Size Estimation (MGSE) performs an estimation of a genome size based on a read mapping to an existing genome sequence assembly. Please find the script and detailed instructions in the MGSE repository. Additional details are described in the corresponding publication 'Mapping-based genome size estimation'.
Prediction of the functional consequences of sequence variants listed in a VCF file. Please find scripts and detailed instructions in the NAVIP repository. We are currently working to make NAVIP available through our web server. Please find additional details in the corresponding publication 'Influence of neighboring small sequence variants on functional impact prediction'.
Material used in this course can be found here.

Material used in this course can be found here. Scripts and instructions of the bioinformatics component are available here.
Material used in this course can be found here. Scripts and instructions of the bioinformatics component are available here.
PuckerLab is hosting the TU Braunschweig iGEM team 2022.

Please find a full list of our publications on the @PuckerLab website.
Do you have questions or are you interested in a collaboration? Please get in touch via email.