A free and open-source cross-platform DNA Sequencing/Visualization Software for bioinformatics research.
A toolkit for instantly performing sequencing, alignment, distance algorithms on DNA strings and visualizing their structure. It's like GParted but for genome sequencing.
The core goal of the project is to be a good general-purpose toolkit for DNA sequencing/alignment/distance analysis and to bring more open-source attention to bioinformatics by promoting more research software in the industry to be open-source. (Read Goals)
Built with ❤️ at FOSSHack 3.0!
UPDATE: This project won FOSSHack 3.0!
- Goals
- Features
- Installation
- Build From Source
- Web API (OpenAPI Schema)
- GUI
- Citations & Acknowledgments
- Contribution
- License
- Project Progress
- First goal is to make DNArchery a good general-purpose toolkit for DNA sequencing/alignment/distance analysis.
- Most utility bioinformatics software used by the industry are commercial (See List), this project is intended to bring a groundwork into making more utilities in the industry open-source.
- The structure of the project is written in such a way that implementing new functionality like algorithms/visualizations are just plugging new functions to the set of utils already present, this is intended to reduce the barrier of entry in encouraging more open-source contributions. (Read Contribution)
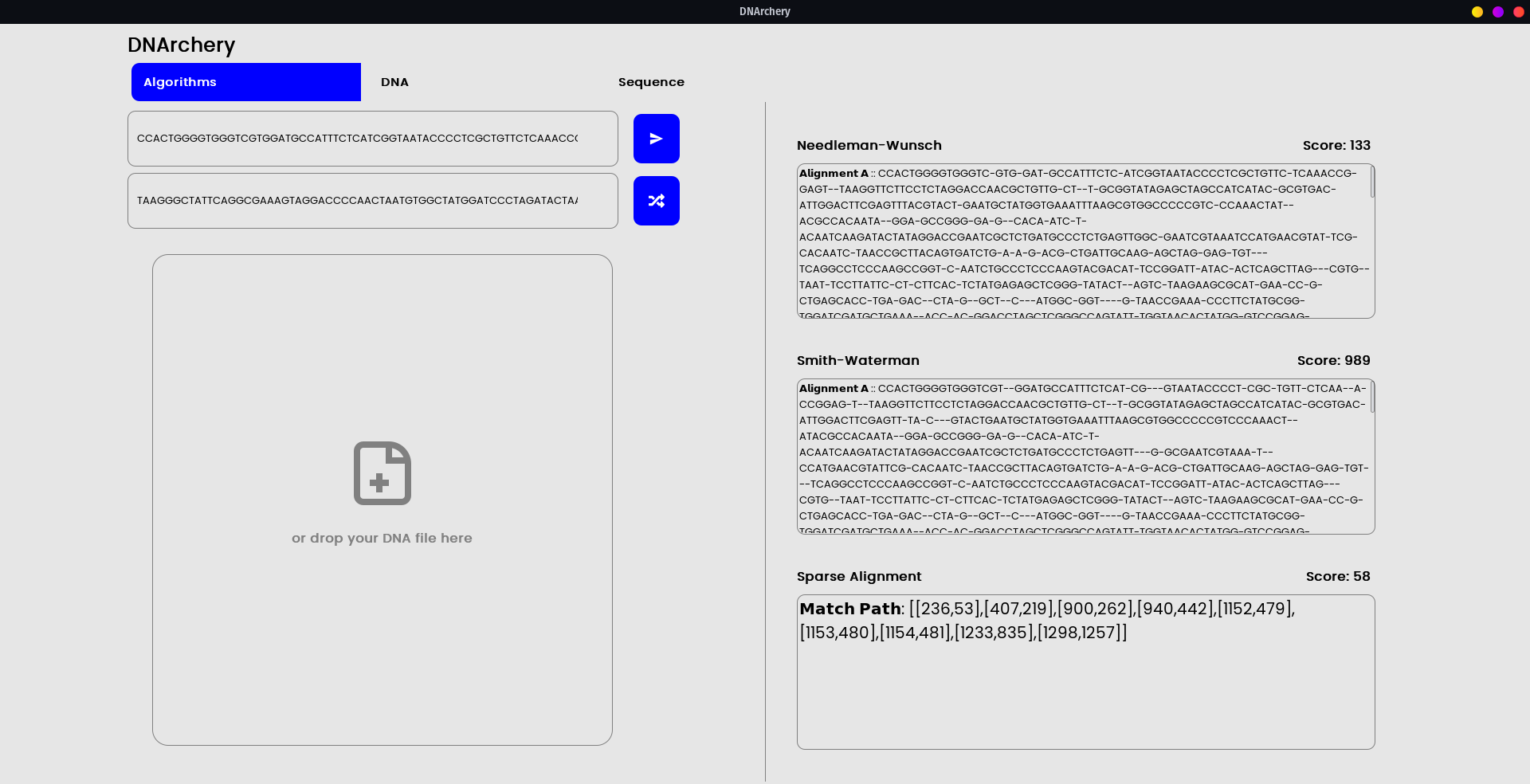
- This tool comprises of an amalgamation of sequencing/alignment/distance algorithms like Needleman–Wunsch algorithm , Smith–Waterman algorithm, and LCSk++ etc. in a single package such that for a general-purpose DNA analysis this could be a go-to utility.
- A secondary goal being, promoting the usage of Rust 🦀 in building more open-source research software as it brings the performance and correctness required for parallel processing like in the case of this utility.
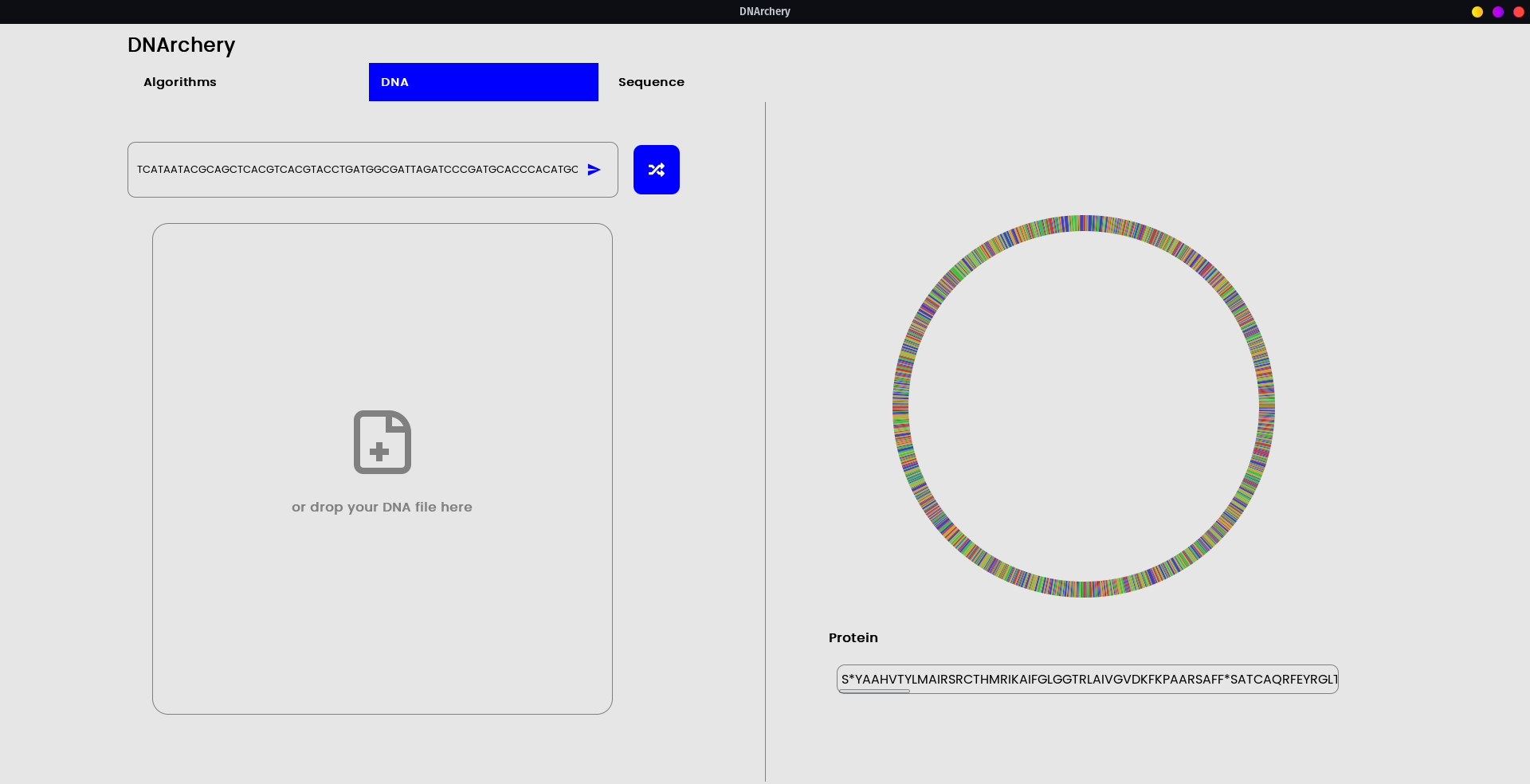
- The core of the utility is exposed as a Webservice API (Rust backend) along with an OpenAPI schema such that UIs or Apps can be built on top, just the like main UI toolkit DNArchery comes with. (See GUI)
- Toolsets:
- Conversions - From DNA to Amino Acids, Proteins, Codon frames, etc.
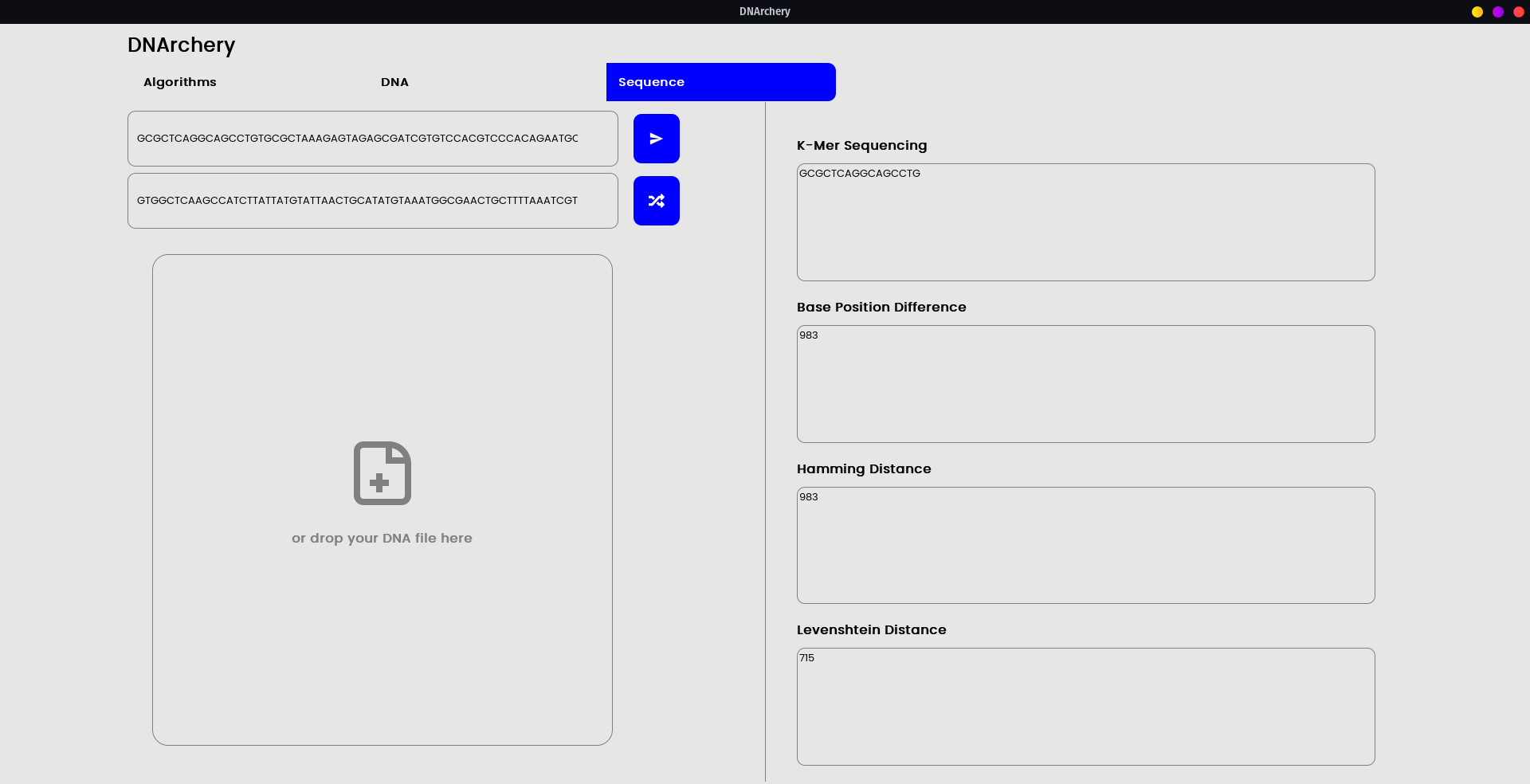
- Sequencing Algorithms - K-mer, Ndiffs, and more.
- Distance/Compare Algorithms - Needleman–Wunsch algorithm , Smith–Waterman algorithm, LCSk++, Hamming distance, Sparse alignments, etc.
- Performance - Processing large FASTA files/DNA sequences are pretty fast as most of them utilize vectorized or parallelized algorithms[example].
Install the executable with the following command:
Note You would need the assets for the GUI integration to work, follow the Build From Source instructions to set up.
$ cargo install --git https://github.com/DNArchery/DNArchery.git
$ dnarcheryPrerequisites:
- Git
- Rust
- Cargo (Automatically installed when installing Rust)
- A C linker (Only for Linux, generally comes pre-installed)
$ git clone https://github.com/DNArchery/DNArchery.git
$ cd DNArchery/
$ # < GUI dependencies >
$ sudo apt-get install -y gir1.2-javascriptcoregtk-4.0
$ sudo apt-get install -y libwebkit2gtk-4.1-dev
$ # </ GUI dependencies >
$ cargo build --release
$ ./target/release/dnarcheryThe first command clones this repository into your local machine and the last two commands enters the directory and builds the source in release mode.
You can see the OpenAPI Schema (Swagger UI) at http://127.0.0.1:1337/swagger-ui/.
 |
The integrated GUI is a proof-of-concept into how the underlying API schema can be utilized.
 |
 |
 |
As stated in the goals section, one of our primary goal is to provide a low barrier contributing opportunity to the bioinformatics open-source space. If you want to add more DNA sequencing/alignment/conversion algorithms, you can browse to src -> core -> and chose which part you want to extend. Every super module in the tree have the same structure, a utils.rs file which contain all functions, you can add a new function and implement it as an exposed actix-web endpoint in the src -> api -> endpoints.rs and that's it.
Ways to contribute:
- Suggest a feature
- Report a bug
- Fix something and open a pull request
- Help me document the code
- Spread the word
Licensed under the MIT License, see LICENSE for more information.
This project wouldn't exist without these resources (libraries/blogs):
FOSSHack Questionnaire:
Q. What was the initial stage of the project?
The idea of the project is to create a utility box of various DNA sequencing algorithms exposed via a API schema such that UIs, Apps can be built on top. The integratred GUI is an example of this.
The initial stage is just a code structure that provides an easy way to embed new algorithms just by adding a new function to one of the
utils.rsfiles. (Either indna/sequence/fasta)This is then exposed via a Web API (Rust backend) (See Code). This API is utilized for the integrated GUI app.
Q. What stage is it in now?
The Rust backend is complete. The GUI on top is a proof-of-concept stage.
Q. How did you get there?
Authors of the project are interested in bioinformatics, during the course of development of this project, we researched about the basic needs in a DNA sequencing software and looked into resources to implement it in code. Thanks to the resources (See Citations & Acknowledgments for Credits), we were able to utilize some libraries for algorithms and we implemented the rest with the documentation.
We chose Rust as the primary language as that's what we are comfortable with and it provides the necessary performance in running compute intensive algorithms.
Q. What is working/not working?
The backend is stable and works according to the specified schema. The GUI is just a wrapper around it that does I/O from the same API.
