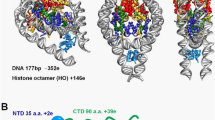
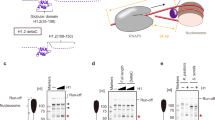
Abstract
It is essential to identify the exact location of the linker histone within nucleosomes, the fundamental packing units of chromatin, in order to understand how condensed, transcriptionally inactive chromatin forms. Here, using a site-specific protein–DNA photo-crosslinking method1, we map the binding site and the orientation of the globular domain of linker histone H5 on mixed-sequence chicken nucleosomes. We show, in contrast to an earlier model2, that the globular domain forms a bridge between one terminus of chromatosomal DNA and the DNA in the vicinity of the dyad axis of symmetry of the core particle. Helix III of the globular domain binds in the major groove of the first helical turn of the chromatosomal DNA, whereas the secondary DNA-binding site on the opposite face of the globular domain of histone H5 makes contact with the nucleosomal DNA close to its midpoint. We also infer that helix I and helix II of the globular domain of histone H5 probably face, respectively, the solvent and the nucleosome. This location places the basic carboxy-terminal region of the globular domain in a position from which it could simultaneously bind the nucleosome-linking DNA strands that exit and enter the nucleosome.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Pendergast, P. S., Chen, Y., Ebright, Y. W. & Ebright R. H. Determination of the orientation of a DNA binding motif in a protein–DNA complex by photocrosslinking. Proc. Natl Acad. Sci. USA> 89 10287–10291 (1992).
Pruss, D.et al. An asymmetric model for the nucleosome: a binding site for linker histones inside the DNA gyres. Science 274, 614–617 (1996).
Thoma, F., Koller, T. & Klug, A. Involvement of histone H1 in the organisation of the nucleosome and of the salt dependent superstructure of chromatin. J. Cell Biol. 83, 403–427 (1979).
Simpson, R. Structure of the chromatosome, a chromatin particle containing 160 bp of DNA and all histones. Biochemistry 17, 5524–5531 (1978).
Allan, J., Crane-Robinson, C. & Aviles, F. X. The structure of histone H1 and its location in chromatin. Nature 288, 675–679 (1980).
Ramakrishnan, V., Finch, J. T., Graziano, V., Lee, P. L. & Sweet, R. M. Crystal structure of the globular domain of histone H5 and its implications for nucleosome binding. Nature 362, 219–223 (1993).
Cerf, C. et al. Homo- and heteronuclear two-dimensional NMR studies of the globular domain of histone H1: full assignment, tertiary structure and comparison with the globular domain of histone H5. Biochemistry 33, 11079–11086 (1994).
Clark, K. L., Halay, E. D., Lai, E. & Burley, S. K. Co-crystal structure of the HNF-3/Fork head DNA-recognition motif resembles histone H5. Nature 364, 412–420 (1993).
Goytisolo, F. A. et al. Identification of two DNA-binding sites on the globular domain of histone H5. EMBO J. 15, 3421–3429 (1996).
Crane-Robinson, C. Where is the globular domain of linker histone located on the nucleosome? Trends Biochem. Sci. 22, 75–77 (1997).
Lambert, S. et al. Neutron scattering studies of chromatosomes. Biochem. Biophys. Res. Commun. 179, 810–816 (1991).
Muyldermans, S. & Travers, A. A. DNA sequence organization in chromatosomes. J. Mol. Biol. 235, 855–870 (1994).
Travers, A. A. & Muyldermans, S. ADNA sequence for positioning chromatosomes. J. Mol. Biol. 257, 486–491 (1996).
Hayes, J. J. Site-directed cleavage of DNA by a linker histone–Fe(II) EDTA conjugate: localization of a globular domain binding site within a nucleosome. Biochemistry 35, 11931–11937 (1996).
Segers, A., Muyldermans, S. & Wyns, L. The interaction of histone H5 and its globular domain with core particles, depleted chromatosomes, polynucleosomes, and a DNA decamer. J. Biol. Chem. 266, 1502–1508 (1991).
Lutter, L. C. Precise location of DNase I cutting sites on the nucleosome core determined by high resolution gel electrophoresis. Nucleic Acids Res. 6, 41–56 (1979).
Hayes, J. J. & Wolffe, A. P. Preferential and asymmetric interaction of linker histones with 5S DNA in the nucleosome. Proc. Natl Acad. Sci. USA 90, 6415–6419 (1993).
Thomas, J. O. & Wilson, C. M. Selective radiolabeling and identification of a strong nucleosome binding site on the globular domain of histone H5. EMBO J. 5, 3531–3537 (1986).
Luger, K., Mäder, A. W., Richmond, R. K., Sargent, D. F. & Richmond, T. J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260 (1997).
An, W., van Holde, K. & Zlatanova, J. Linker histone protection of chromatosomes reconstituted on 5S rDNA from Xenopus borealis: a reinvestigation. Nucleic Acids Res. (in the press).
Thomas, J. O., Rees, C. & Finch, J. T. Cooperative binding of the globular domains of histones H1 and H5 to DNA. Nucleic Acids Res. 20, 187–194 (1992).
Drwe, H. R. & McCall, M. J. Structural analysis of a reconstituted DNA containing three histone octamers and histone H5. J. Mol. Biol. 197, 485–511 (1987).
Staynov, D. Z. & Crane-Robinson, C. Footprinting of linker histones H5 and H1 on the nucleosome. EMBO J. 7, 3685–3691 (1988).
Pehrson, J. R. Thymine dimer formation as a probe of the path of DNA in and between nucleosomes in intact chromatin. Proc. Natl Acad. Sci. USA 86, 9149–9153 (1989).
Mirzabekov, A., Pruss, D. & Ebralidse, K. K. Chromatin superstructure dependent crosslinking with DNA of the histone H5 residues Thr1, His25 and His62. J. Mol. Biol. 211, 479–491 (1989).
Hamiche, A., Schulz, P., Ramakrishnan, V., Oudet, P. & Prunell, A. Linker histone dependent DNA structure in mononucleosomes. J. Mol. Biol. 257, 30–42 (1996).
Hayes, J. J., Pruss, D. & Wolffe, A. P. Contacts of the globular domain of histone H5 and core histones with DNA in a “chromatosome”. Proc. Natl Acad. Sci. USA 91, 7817–7821 (1994).
Howe, L. & Ausió, J. Nucleosome translational position, not histone acetylation, determines TFIIIA binding to nucleosomes Xenopus laevis rRNA genes. Mol. Cell. Biol. 18, 1156–1162 (1998).
Pannetta, G., Buttinelli, M., Flaus, A., Richmond, T. J. & Rhodes, D. Differential nucleosome positioning on Xenopus oocyte and somatic 5S RNA genes determines both RFIIA and H1 binding: a mechanism for selective H1 repression. J. Mol. Biol. (in the press).
Acknowledgements
We thank T. Hamelryck and Y. Geunes for help with figures, and L. Wyns for helpful discussions. Z.Y.B. received a grant from the VlAB. V.R. was supported by a Public Health Service grant from the NIH. This work was supported by grants from (N)FWO and the OZR-VUB.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhou, YB., Gerchman, S., Ramakrishnan, V. et al. Position and orientation of the globular domain of linker histone H5 on the nucleosome. Nature 395, 402–405 (1998). https://doi.org/10.1038/26521
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/26521