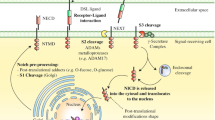
Abstract
The ubiquitin proteasome system (UPS) is a highly conserved way to regulate protein turnover in cells. The UPS hydrolyzes and destroys variant or misfolded proteins and finely regulates proteins involved in differentiation, apoptosis, and other biological processes. This system is a key regulatory factor in the proliferation, differentiation, and collagen secretion of skin fibroblasts. E3 ubiquitin protein ligases Parkin and NEDD4 regulate multiple signaling pathways in keloid. Tumor necrosis factor (TNF) receptor-associated factor 4 (TRAF4) binding with deubiquitinase USP10 can induce p53 destabilization and promote keloid-derived fibroblast proliferation. The UPS participates in the occurrence and development of hypertrophic scars by regulating the transforming growth factor (TGF)-β/Smad signaling pathway. An initial study suggests that TNFα-induced protein 3 (TNFAIP3) polymorphisms may be significantly associated with scleroderma susceptibility in individuals of Caucasian descent. Sumoylation and multiple ubiquitin ligases, including Smurfs, UFD2, and KLHL42, play vital roles in scleroderma by targeting the TGF-β/Smad signaling pathway. In the future, drugs targeting E3 ligases and deubiquitinating enzymes have great potential for the treatment of skin fibrosis.



Similar content being viewed by others
References
Zhao X, Psarianos P, Ghoraie LS, Yip K, Goldstein D, Gilbert R, et al. Metabolic regulation of dermal fibroblasts contributes to skin extracellular matrix homeostasis and fibrosis. Nat Metab. 2019;1(1):147–57. https://doi.org/10.1038/s42255-018-0008-5.
Griffin MF, des Jardins-Park HE, Mascharak S, Borrelli MR, Longaker MT. Understanding the impact of fibroblast heterogeneity on skin fibrosis. Dis Models Mech. 2020. https://doi.org/10.1242/dmm.044164.
Do NN, Eming SA. Skin fibrosis: models and mechanisms. Curr Res Transl Med. 2016;64(4):185–93. https://doi.org/10.1016/j.retram.2016.06.003.
Li Y, Zhang J, Lei Y, Lyu L, Zuo R, Chen T. MicroRNA-21 in skin fibrosis: potential for diagnosis and treatment. Mol Diagn Ther. 2017;21(6):633–42. https://doi.org/10.1007/s40291-017-0294-8.
Coentro JQ, Pugliese E, Hanley G, Raghunath M, Zeugolis DI. Current and upcoming therapies to modulate skin scarring and fibrosis. Adv Drug Deliv Rev. 2019;146:37–59. https://doi.org/10.1016/j.addr.2018.08.009.
Moon SJ, Bae JM, Park KS, Tagkopoulos I, Kim KJ. Compendium of skin molecular signatures identifies key pathological features associated with fibrosis in systemic sclerosis. Ann Rheum Dis. 2019;78(6):817–25. https://doi.org/10.1136/annrheumdis-2018-214778.
Schulz JN, Plomann M, Sengle G, Gullberg D, Krieg T, Eckes B. New developments on skin fibrosis—essential signals emanating from the extracellular matrix for the control of myofibroblasts. Matrix Biol. 2018;68–69:522–32. https://doi.org/10.1016/j.matbio.2018.01.025.
Wei J, Melichian D, Komura K, Hinchcliff M, Lam AP, Lafyatis R, et al. Canonical Wnt signaling induces skin fibrosis and subcutaneous lipoatrophy: a novel mouse model for scleroderma? Arthritis Rheum. 2011;63(6):1707–17. https://doi.org/10.1002/art.30312.
Akhmetshina A, Palumbo K, Dees C, Bergmann C, Venalis P, Zerr P, et al. Activation of canonical Wnt signalling is required for TGF-beta-mediated fibrosis. Nat Commun. 2012;3:735. https://doi.org/10.1038/ncomms1734.
Lear TB, Lockwood KC, Larsen M, Tuncer F, Kennerdell JR, Morse C, et al. Kelch-like protein 42 is a profibrotic ubiquitin E3 ligase involved in systemic sclerosis. J Biol Chem. 2020;295(13):4171–80. https://doi.org/10.1074/jbc.AC119.012066.
Mori Y, Chen SJ, Varga J. Modulation of endogenous Smad expression in normal skin fibroblasts by transforming growth factor-beta. Exp Cell Res. 2000;258(2):374–83. https://doi.org/10.1006/excr.2000.4930.
Galant C, Marchandise J, Stoenoiu MS, Ducreux J, De Groof A, Pirenne S, et al. Overexpression of ubiquitin-specific peptidase 15 in systemic sclerosis fibroblasts increases response to transforming growth factor beta. Rheumatology. 2019;58(4):708–18. https://doi.org/10.1093/rheumatology/key401.
Radovanac K, Morgner J, Schulz JN, Blumbach K, Patterson C, Geiger T, et al. Stabilization of integrin-linked kinase by the Hsp90-CHIP axis impacts cellular force generation, migration and the fibrotic response. EMBO J. 2013;32(10):1409–24. https://doi.org/10.1038/emboj.2013.90.
Roque W, Summer R, Romero F. Fine-tuning the ubiquitin-proteasome system to treat pulmonary fibrosis. Connect Tissue Res. 2019;60(1):50–61. https://doi.org/10.1080/03008207.2018.1529174.
Wang Y, Le WD. Autophagy and ubiquitin-proteasome system. Adv Exp Med Biol. 2019;1206:527–50. https://doi.org/10.1007/978-981-15-0602-4_25.
Bax M, McKenna J, Do-Ha D, Stevens CH, Higginbottom S, Balez R, et al. The ubiquitin proteasome system is a key regulator of pluripotent stem cell survival and motor neuron differentiation. Cells. 2019. https://doi.org/10.3390/cells8060581.
Iborra RT, Machado-Lima A, Okuda LS, Pinto PR, Nakandakare ER, Machado UF, et al. AGE-albumin enhances ABCA1 degradation by ubiquitin-proteasome and lysosomal pathways in macrophages. J Diabetes Complicat. 2018;32(1):1–10. https://doi.org/10.1016/j.jdiacomp.2017.09.012.
Li M, Rong Y, Chuang YS, Peng D, Emr SD. Ubiquitin-dependent lysosomal membrane protein sorting and degradation. Mol Cell. 2015;57(3):467–78. https://doi.org/10.1016/j.molcel.2014.12.012.
Zhao Y, Brickner JR, Majid MC, Mosammaparast N. Crosstalk between ubiquitin and other post-translational modifications on chromatin during double-strand break repair. Trends Cell Biol. 2014;24(7):426–34. https://doi.org/10.1016/j.tcb.2014.01.005.
Amm I, Sommer T, Wolf DH. Protein quality control and elimination of protein waste: the role of the ubiquitin-proteasome system. Biochem Biophys Acta. 2014;1843(1):182–96. https://doi.org/10.1016/j.bbamcr.2013.06.031.
de Jong A, Merkx R, Berlin I, Rodenko B, Wijdeven RH, El Atmioui D, et al. Ubiquitin-based probes prepared by total synthesis to profile the activity of deubiquitinating enzymes. ChemBioChem. 2012;13(15):2251–8. https://doi.org/10.1002/cbic.201200497.
Gong P, Davidson GA, Gui W, Yang K, Bozza WP, Zhuang Z. Activity-based ubiquitin-protein probes reveal target protein specificity of deubiquitinating enzymes. Chem Sci. 2018;9(40):7859–65. https://doi.org/10.1039/c8sc01573b.
Yamanaka S, Sato Y, Oikawa D, Goto E, Fukai S, Tokunaga F, et al. Subquinocin, a small molecule inhibitor of CYLD and USP-family deubiquitinating enzymes, promotes NF-kappaB signaling. Biochem Biophys Res Commun. 2020;524(1):1–7. https://doi.org/10.1016/j.bbrc.2019.12.049.
Xu C, Peng Y, Zhang Q, Xu XP, Kong XM, Shi WF. USP4 positively regulates RLR-induced NF-kappaB activation by targeting TRAF6 for K48-linked deubiquitination and inhibits enterovirus 71 replication. Sci Rep. 2018;8(1):13418. https://doi.org/10.1038/s41598-018-31734-6.
Li Z, Hao Q, Luo J, Xiong J, Zhang S, Wang T, et al. USP4 inhibits p53 and NF-kappaB through deubiquitinating and stabilizing HDAC2. Oncogene. 2016;35(22):2902–12. https://doi.org/10.1038/onc.2015.349.
Fang G, Fu Y, Li S, Qiu J, Kuang M, Lin S, et al. The USP14-NLRC5 pathway inhibits titanium particle-induced osteolysis in mice by suppressing NF-kappaB and PI3K/AKT activities. J Biol Chem. 2020;295(20):7018–32. https://doi.org/10.1074/jbc.RA119.012495.
Huang K, Zhao X. USP9X prevents AGEs-induced upregulation of FN and TGF-beta1 through activating Nrf2-ARE pathway in rat glomerular mesangial cells. Exp Cell Res. 2020;393(2):112100. https://doi.org/10.1016/j.yexcr.2020.112100.
Istomine R, Alvarez F, Almadani Y, Philip A, Piccirillo CA. The deubiquitinating enzyme ubiquitin-specific peptidase 11 potentiates TGF-beta signaling in CD4(+) T cells to facilitate Foxp3(+) regulatory T and TH17 cell differentiation. J Immunol. 2019;203(9):2388–400. https://doi.org/10.4049/jimmunol.1801689.
Chou CK, Chang YT, Korinek M, Chen YT, Yang YT, Leu S, et al. The regulations of deubiquitinase USP15 and its pathophysiological mechanisms in diseases. Int J Mol Sci. 2017;18:3. https://doi.org/10.3390/ijms18030483.
Stanley-Hasnain S, Hauck L, Grothe D, Aschar-Sobbi R, Beca S, Butany J, et al. p53 and Mdm2 act synergistically to maintain cardiac homeostasis and mediate cardiomyocyte cell cycle arrest through a network of microRNAs. Cell Cycle. 2017;16(17):1585–600. https://doi.org/10.1080/15384101.2017.1346758.
Zhang X, Berger FG, Yang J, Lu X. USP4 inhibits p53 through deubiquitinating and stabilizing ARF-BP1. EMBO J. 2011;30(11):2177–89. https://doi.org/10.1038/emboj.2011.125.
Dayal S, Sparks A, Jacob J, Allende-Vega N, Lane DP, Saville MK. Suppression of the deubiquitinating enzyme USP5 causes the accumulation of unanchored polyubiquitin and the activation of p53. J Biol Chem. 2009;284(8):5030–41. https://doi.org/10.1074/jbc.M805871200.
Meiners S, Evankovich J, Mallampalli RK. The ubiquitin proteasome system as a potential therapeutic target for systemic sclerosis. Transl Res. 2018;198:17–28. https://doi.org/10.1016/j.trsl.2018.03.003.
Wang W, Luo J, Sheng W, Xue J, Li M, Ji J, et al. Proteomic profiling of radiation-induced skin fibrosis in rats: targeting the ubiquitin-proteasome system. Int J Radiat Oncol Biol Phys. 2016;95(2):751–60. https://doi.org/10.1016/j.ijrobp.2016.01.021.
Li WB, Liu S, Zhang MZ, Liu H, Dong XH, Hao Y, et al. Hyperbaric oxygen therapy relieved pruritus and pain of keloid patients. Am J Transl Res. 2020;12(2):574–82.
Tsai CH, Ogawa R. Keloid research: current status and future directions. Scars Burns Heal. 2019;5:2059513119868659. https://doi.org/10.1177/2059513119868659.
Li Q, Qin Z, Nie F, Bi H, Zhao R, Pan B, et al. Metabolic reprogramming in keloid fibroblasts: aerobic glycolysis and a novel therapeutic strategy. Biochem Biophys Res Commun. 2018;496(2):641–7. https://doi.org/10.1016/j.bbrc.2018.01.068.
Deschene K, Celeste C, Boerboom D, Theoret CL. Hypoxia regulates the expression of extracellular matrix associated proteins in equine dermal fibroblasts via HIF1. J Dermatol Sci. 2012;65(1):12–8. https://doi.org/10.1016/j.jdermsci.2011.09.006.
Jusman SWA, Sari DH, Ningsih SS, Hardiany NS, Sadikin M. Role of hypoxia inducible factor-1 alpha (HIF-1alpha) in cytoglobin expression and fibroblast proliferation of keloids. Kobe J Med Sci. 2019;65(1):E10–8.
Lin X, Wang Y, Jiang Y, Xu M, Pang Q, Sun J, et al. Sumoylation enhances the activity of the TGF-beta/SMAD and HIF-1 signaling pathways in keloids. Life Sci. 2020;255:117859. https://doi.org/10.1016/j.lfs.2020.117859.
Elswood J, Pearson SJ, Payne HR, Barhoumi R, Rijnkels M, Porter WW. Autophagy regulates functional differentiation of mammary epithelial cells. Autophagy. 2020. https://doi.org/10.1080/15548627.2020.1720427.
Lai HH, Li JN, Wang MY, Huang HY, Croce CM, Sun HL, et al. HIF-1alpha promotes autophagic proteolysis of Dicer and enhances tumor metastasis. J Clin Investig. 2018;128(2):625–43. https://doi.org/10.1172/JCI89212.
Lei R, Shen J, Zhang S, Liu A, Chen X, Wang Y, et al. Inactivating the ubiquitin ligase Parkin suppresses cell proliferation and induces apoptosis in human keloids. J Cell Physiol. 2019. https://doi.org/10.1002/jcp.28332.
Deng CC, Zhu DH, Chen YJ, Huang TY, Peng Y, Liu SY, et al. TRAF4 promotes fibroblast proliferation in keloids by destabilizing p53 via interacting with the deubiquitinase USP10. J Investig Dermatol. 2019;139(9):1925 e5-1935. https://doi.org/10.1016/j.jid.2019.03.1136.
Kim E, Kim W, Lee S, Chun J, Kang J, Park G, et al. TRAF4 promotes lung cancer aggressiveness by modulating tumor microenvironment in normal fibroblasts. Sci Rep. 2017;7(1):8923. https://doi.org/10.1038/s41598-017-09447-z.
Zhang Z, Guo M, Shen M, Kong D, Zhang F, Shao J, et al. The BRD7-P53-SLC25A28 axis regulates ferroptosis in hepatic stellate cells. Redox Biol. 2020;36:101619. https://doi.org/10.1016/j.redox.2020.101619.
Mijit M, Caracciolo V, Melillo A, Amicarelli F, Giordano A. Role of p53 in the regulation of cellular senescence. Biomolecules. 2020. https://doi.org/10.3390/biom10030420.
El-Mai M, Marzullo M, de Castro IP, Ferreira MG. Opposing p53 and mTOR/AKT promote an in vivo switch from apoptosis to senescence upon telomere shortening in zebrafish. eLife. 2020. https://doi.org/10.7554/eLife.54935.
Liu J, Xia H, Kim M, Xu L, Li Y, Zhang L, et al. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell. 2011;147(1):223–34. https://doi.org/10.1016/j.cell.2011.08.037.
Kumar S, Brown A, Tchounwou PB. Trisenox disrupts MDM2-DAXX-HAUSP complex and activates p53, cell cycle regulation and apoptosis in acute leukemia cells. Oncotarget. 2018;9(69):33138–48. https://doi.org/10.18632/oncotarget.26025.
Liu J, Chung HJ, Vogt M, Jin Y, Malide D, He L, et al. JTV1 co-activates FBP to induce USP29 transcription and stabilize p53 in response to oxidative stress. EMBO J. 2011;30(5):846–58. https://doi.org/10.1038/emboj.2011.11.
Hock AK, Vigneron AM, Carter S, Ludwig RL, Vousden KH. Regulation of p53 stability and function by the deubiquitinating enzyme USP42. EMBO J. 2011;30(24):4921–30. https://doi.org/10.1038/emboj.2011.419.
Hu C, Zhang M, Moses N, Hu CL, Polin L, Chen W, et al. The USP10-HDAC6 axis confers cisplatin resistance in non-small cell lung cancer lacking wild-type p53. Cell Death Dis. 2020;11(5):328. https://doi.org/10.1038/s41419-020-2519-8.
Yuan J, Luo K, Zhang L, Cheville JC, Lou Z. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell. 2010;140(3):384–96. https://doi.org/10.1016/j.cell.2009.12.032.
Fujita M, Yamamoto Y, Jiang JJ, Atsumi T, Tanaka Y, Ohki T, et al. NEDD4 is involved in inflammation development during keloid formation. J Invest Dermatol. 2019;139(2):333–41. https://doi.org/10.1016/j.jid.2018.07.044.
Scott JL, Frick CT, Johnson KA, Liu H, Yong SS, Varney AG, et al. Molecular analysis of membrane targeting by the C2 domain of the E3 ubiquitin ligase Smurf1. Biomolecules. 2020. https://doi.org/10.3390/biom10020229.
Jiang H, Thomas SN, Chen Z, Chiang CY, Cole PA. Comparative analysis of the catalytic regulation of NEDD4-1 and WWP2 ubiquitin ligases. J Biol Chem. 2019;294(46):17421–36. https://doi.org/10.1074/jbc.RA119.009211.
Marneros AG. A role for the E3 ubiquitin ligase NEDD4 in keloid pathogenesis. J Invest Dermatol. 2019;139(2):279–80. https://doi.org/10.1016/j.jid.2018.09.007.
Chung S, Nakashima M, Zembutsu H, Nakamura Y. Possible involvement of NEDD4 in keloid formation; its critical role in fibroblast proliferation and collagen production. Proc Jpn Acad Ser B Phys Biol Sci. 2011;87(8):563–73. https://doi.org/10.2183/pjab.87.563.
Zhao Y, Liu SL, Xie J, Ding MQ, Lu MZ, Zhang LF, et al. NEDD4 single nucleotide polymorphism rs2271289 is associated with keloids in Chinese Han population. Am J Transl Res. 2016;8(2):544–55.
Ogawa R, Watanabe A, Than Naing B, Sasaki M, Fujita A, Akaishi S, et al. Associations between keloid severity and single-nucleotide polymorphisms: importance of rs8032158 as a biomarker of keloid severity. J Invest Dermatol. 2014;134(7):2041–3. https://doi.org/10.1038/jid.2014.71.
Nakashima M, Chung S, Takahashi A, Kamatani N, Kawaguchi T, Tsunoda T, et al. A genome-wide association study identifies four susceptibility loci for keloid in the Japanese population. Nat Genet. 2010;42(9):768–71. https://doi.org/10.1038/ng.645.
Zhu F, Wu B, Li P, Wang J, Tang H, Liu Y, et al. Association study confirmed susceptibility loci with keloid in the Chinese Han population. PLoS ONE. 2013;8(5):e62377. https://doi.org/10.1371/journal.pone.0062377.
Wang Y, Yuan B, Qiao L, Yang H, Li X. STAT3 operates as a novel transcription factor that regulates NEDD4 in Keloid. Biochem Biophys Res Commun. 2019;518(4):638–43. https://doi.org/10.1016/j.bbrc.2019.08.110.
Ren X, Duan L, He Q, Zhang Z, Zhou Y, Wu D, et al. Identification of niclosamide as a new small-molecule inhibitor of the STAT3 signaling pathway. ACS Med Chem Lett. 2010;1(9):454–9. https://doi.org/10.1021/ml100146z.
Sang PF, Wang H, Wang M, Hu C, Zhang JS, Li XJ, et al. NEDD4-1 and PTEN expression in keloid scarring. Genet Mol Res. 2015;14(4):13467–75. https://doi.org/10.4238/2015.October.28.7.
Zhang J, Li Y, Bai X, Li Y, Shi J, Hu D. Recent advances in hypertrophic scar. Histol Histopathol. 2018;33(1):27–39. https://doi.org/10.14670/HH-11-908.
Hesketh M, Sahin KB, West ZE, Murray RZ. Macrophage phenotypes regulate scar formation and chronic wound healing. Int J Mol Sci. 2017. https://doi.org/10.3390/ijms18071545.
Zhu Z, Ding J, Ma Z, Iwashina T, Tredget EE. Systemic depletion of macrophages in the subacute phase of wound healing reduces hypertrophic scar formation. Wound Repair Regener. 2016;24(4):644–56. https://doi.org/10.1111/wrr.12442.
Hwangbo C, Tae N, Lee S, Kim O, Park OK, Kim J, et al. Syntenin regulates TGF-beta1-induced Smad activation and the epithelial-to-mesenchymal transition by inhibiting caveolin-mediated TGF-beta type I receptor internalization. Oncogene. 2016;35(3):389–401. https://doi.org/10.1038/onc.2015.100.
Jung H, Seong HA, Manoharan R, Ha H. Serine-threonine kinase receptor-associated protein inhibits apoptosis signal-regulating kinase 1 function through direct interaction. J Biol Chem. 2010;285(1):54–70. https://doi.org/10.1074/jbc.M109.045229.
Zhang J, Na S, Pan S, Long S, Xin Y, Jiang Q, et al. Inhibition of USP4 attenuates pathological scarring by downregulation of the TGFbeta/Smad signaling pathway. Mol Med Rep. 2019;20(2):1429–35. https://doi.org/10.3892/mmr.2019.10370.
Fukui S, Nagasaka K, Miyagawa Y, Kikuchi-Koike R, Kawata Y, Kanda R, et al. The proteasome deubiquitinase inhibitor bAP15 downregulates TGF-beta/Smad signaling and induces apoptosis via UCHL5 inhibition in ovarian cancer. Oncotarget. 2019;10(57):5932–48. https://doi.org/10.18632/oncotarget.27219.
Huang Y, Wang Y, Wang X, Lin L, Wang P, Sun J, et al. The Effects of the transforming growth factor-beta1 (TGF-beta1) signaling pathway on cell proliferation and cell migration are mediated by ubiquitin specific protease 4 (USP4) in hypertrophic scar tissue and primary fibroblast cultures. Med Sci Monit. 2020;26:e920736. https://doi.org/10.12659/MSM.920736.
Inoue Y, Imamura T. Regulation of TGF-beta family signaling by E3 ubiquitin ligases. Cancer Sci. 2008;99(11):2107–12. https://doi.org/10.1111/j.1349-7006.2008.00925.x.
Koganti P, Levy-Cohen G, Blank M. Smurfs in protein homeostasis, signaling, and cancer. Front Oncol. 2018;8:295. https://doi.org/10.3389/fonc.2018.00295.
David D, Nair SA, Pillai MR. Smurf E3 ubiquitin ligases at the cross roads of oncogenesis and tumor suppression. Biochem Biophys Acta. 2013;1835(1):119–28. https://doi.org/10.1016/j.bbcan.2012.11.003.
Malonis RJ, Fu W, Jelcic MJ, Thompson M, Canter BS, Tsikitis M, et al. RNF11 sequestration of the E3 ligase SMURF2 on membranes antagonizes SMAD7 down-regulation of transforming growth factor beta signaling. J Biol Chem. 2017;292(18):7435–51. https://doi.org/10.1074/jbc.M117.783662.
Zhang Z, Kuang F, Liu CL, Chen B, Tang WB, Li XJ. Effects of silencing Smad ubiquitination regulatory factor 2 on the function of human hypertrophic scar-derived fibroblasts. Zhonghua shao shang za zhi Zhonghua shaoshang zazhi Chin J Burns. 2017;33(3):145–51. https://doi.org/10.3760/cma.j.issn.1009-2587.2017.03.004.
Zhang Z, Finnerty CC, He J, Herndon DN. Smad ubiquitination regulatory factor 2 expression is enhanced in hypertrophic scar fibroblasts from burned children. Burns. 2012;38(2):236–46. https://doi.org/10.1016/j.burns.2011.08.012.
Tecalco-Cruz AC, Rios-Lopez DG, Vazquez-Victorio G, Rosales-Alvarez RE, Macias-Silva M. Transcriptional cofactors Ski and SnoN are major regulators of the TGF-beta/Smad signaling pathway in health and disease. Signal Transduct Target Ther. 2018;3:15. https://doi.org/10.1038/s41392-018-0015-8.
Makino Y, Yoon JH, Bae E, Kato M, Miyazawa K, Ohira T, et al. Repression of Smad3 by Stat3 and c-Ski/SnoN induces gefitinib resistance in lung adenocarcinoma. Biochem Biophys Res Commun. 2017;484(2):269–77. https://doi.org/10.1016/j.bbrc.2017.01.093.
Wang Y, Liu L, Peng W, Liu H, Liang L, Zhang X, et al. Ski-related novel protein suppresses the development of diabetic nephropathy by modulating transforming growth factor-beta signaling and microRNA-21 expression. J Cell Physiol. 2019;234(10):17925–36. https://doi.org/10.1002/jcp.28425.
Xu H, Sun F, Li X, Sun L. Down-regulation of miR-23a inhibits high glucose-induced EMT and renal fibrogenesis by up-regulation of SnoN. Hum Cell. 2018;31(1):22–32. https://doi.org/10.1007/s13577-017-0180-z.
Liu L, Shi M, Wang Y, Zhang C, Su B, Xiao Y, et al. SnoN upregulation ameliorates renal fibrosis in diabetic nephropathy. PLoS ONE. 2017;12(3):e0174471. https://doi.org/10.1371/journal.pone.0174471.
Zeglinski MR, Hnatowich M, Jassal DS, Dixon IM. SnoN as a novel negative regulator of TGF-beta/Smad signaling: a target for tailoring organ fibrosis. Am J Physiol Heart Circ Physiol. 2015;308(2):H75-82. https://doi.org/10.1152/ajpheart.00453.2014.
Sun GF, Li HC, Zhan YP, Zhang XF, Pan LY, Chen YF, et al. SnoN residue (1–366) attenuates hypertrophic scars through resistance to transforming growth factor-beta1-induced degradation. Lab Investig. 2019;99(12):1861–73. https://doi.org/10.1038/s41374-019-0302-1.
Li P, Liu P, Xiong RP, Chen XY, Zhao Y, Lu WP, et al. Ski, a modulator of wound healing and scar formation in the rat skin and rabbit ear. J Pathol. 2011;223(5):659–71. https://doi.org/10.1002/path.2831.
Arno AI, Gauglitz GG, Barret JP, Jeschke MG. New molecular medicine-based scar management strategies. Burns. 2014;40(4):539–51. https://doi.org/10.1016/j.burns.2013.11.010.
Moore DF, Kramer E, Eltaraboulsi R, Steen VD. Increased morbidity and mortality of scleroderma in African Americans compared to Non-African Americans. Arthritis Care Res. 2019;71(9):1154–63. https://doi.org/10.1002/acr.23861.
Gordon SM, Hughes JB, Nee R, Stitt RS, Bailey WT, Little DJ, et al. Systemic sclerosis medications and risk of scleroderma renal crisis. BMC Nephrol. 2019;20(1):279. https://doi.org/10.1186/s12882-019-1467-y.
Denton CP, Wells AU, Coghlan JG. Major lung complications of systemic sclerosis. Nat Rev Rheumatol. 2018;14(9):511–27. https://doi.org/10.1038/s41584-018-0062-0.
Bussone G, Mouthon L. Interstitial lung disease in systemic sclerosis. Autoimmun Rev. 2011;10(5):248–55. https://doi.org/10.1016/j.autrev.2010.09.012.
Yang L, Jin L, Ke Y, Fan X, Zhang T, Zhang C, et al. E3 ligase Trim21 ubiquitylates and stabilizes keratin 17 to induce STAT3 activation in psoriasis. J Invest Dermatol. 2018;138(12):2568–77. https://doi.org/10.1016/j.jid.2018.05.016.
Shen J, Yu Z, Li N. The E3 ubiquitin ligase RNF146 promotes colorectal cancer by activating the Wnt/beta-catenin pathway via ubiquitination of Axin1. Biochem Biophys Res Commun. 2018;503(2):991–7. https://doi.org/10.1016/j.bbrc.2018.06.107.
Moren A, Imamura T, Miyazono K, Heldin CH, Moustakas A. Degradation of the tumor suppressor Smad4 by WW and HECT domain ubiquitin ligases. J Biol Chem. 2005;280(23):22115–23. https://doi.org/10.1074/jbc.M414027200.
Long Y, Chen W, Du Q, Zuo X, Zhu H. Ubiquitination in scleroderma fibrosis and its treatment. Front Immunol. 2018;9:2383. https://doi.org/10.3389/fimmu.2018.02383.
Tang Y, Zha L, Zeng X, Yu Z. Identification of biomarkers related to systemic sclerosis with or without pulmonary hypertension using co-expression analysis. J Comput Biol. 2020. https://doi.org/10.1089/cmb.2019.0492.
Gao L, Emond MJ, Louie T, Cheadle C, Berger AE, Rafaels N, et al. Identification of rare variants in ATP8B4 as a risk factor for systemic sclerosis by whole-exome sequencing. Arthritis Rheumatol. 2016;68(1):191–200. https://doi.org/10.1002/art.39449.
Dieude P, Guedj M, Wipff J, Ruiz B, Riemekasten G, Matucci-Cerinic M, et al. Association of the TNFAIP3 rs5029939 variant with systemic sclerosis in the European Caucasian population. Ann Rheum Dis. 2010;69(11):1958–64. https://doi.org/10.1136/ard.2009.127928.
Just S, Nishanth G, Buchbinder JH, Wang X, Naumann M, Lavrik I, et al. A20 curtails primary but augments secondary CD8(+) T cell responses in intracellular bacterial infection. Sci Rep. 2016;6:39796. https://doi.org/10.1038/srep39796.
Duong BH, Onizawa M, Oses-Prieto JA, Advincula R, Burlingame A, Malynn BA, et al. A20 restricts ubiquitination of pro-interleukin-1beta protein complexes and suppresses NLRP3 inflammasome activity. Immunity. 2015;42(1):55–67. https://doi.org/10.1016/j.immuni.2014.12.031.
Nakamura BN, Glazier A, Kattah MG, Duong B, Jia Y, Campo D, et al. A20 regulates canonical wnt-signaling through an interaction with RIPK4. PLoS ONE. 2018;13(5):e0195893. https://doi.org/10.1371/journal.pone.0195893.
Wei P, Yang Y, Guo X, Hei N, Lai S, Assassi S, et al. Identification of an association of TNFAIP3 polymorphisms with matrix metalloproteinase expression in fibroblasts in an integrative study of systemic sclerosis-associated genetic and environmental factors. Arthritis Rheumatol. 2016;68(3):749–60. https://doi.org/10.1002/art.39476.
Koumakis E, Giraud M, Dieude P, Cohignac V, Cuomo G, Airo P, et al. Brief report: candidate gene study in systemic sclerosis identifies a rare and functional variant of the TNFAIP3 locus as a risk factor for polyautoimmunity. Arthritis Rheum. 2012;64(8):2746–52. https://doi.org/10.1002/art.34490.
Meng XM, Chung AC, Lan HY. Role of the TGF-beta/BMP-7/Smad pathways in renal diseases. Clin Sci. 2013;124(4):243–54. https://doi.org/10.1042/CS20120252.
Li TF, Darowish M, Zuscik MJ, Chen D, Schwarz EM, Rosier RN, et al. Smad3-deficient chondrocytes have enhanced BMP signaling and accelerated differentiation. J Bone Miner Res. 2006;21(1):4–16. https://doi.org/10.1359/JBMR.050911.
Chen H, Yang R, Tang Y, Fu X. Effects of curcumin on artery blood gas index of rats with pulmonary fibrosis caused by paraquat poisoning and the expression of Smad 4, Smurf 2, interleukin-4 and interferon-gamma. Exp Ther Med. 2019;17(5):3664–70. https://doi.org/10.3892/etm.2019.7341.
Liu C, van Dyk D, Choe V, Yan J, Majumder S, Costanzo M, et al. Ubiquitin ligase Ufd2 is required for efficient degradation of Mps1 kinase. J Biol Chem. 2011;286(51):43660–7. https://doi.org/10.1074/jbc.M111.286229.
Spinette S, Lengauer C, Mahoney JA, Jallepalli PV, Wang Z, Casciola-Rosen L, et al. Ufd2, a novel autoantigen in scleroderma, regulates sister chromatid separation. Cell Cycle. 2004;3(12):1638–44.
Jacko AM, Nan L, Li S, Tan J, Zhao J, Kass DJ, et al. De-ubiquitinating enzyme, USP11, promotes transforming growth factor beta-1 signaling through stabilization of transforming growth factor beta receptor II. Cell Death Dis. 2016;7(11):e2474. https://doi.org/10.1038/cddis.2016.371.
Khodzhigorova A, Distler A, Lang V, Dees C, Schneider H, Beyer C, et al. Inhibition of sumoylation prevents experimental fibrosis. Ann Rheum Dis. 2012;71(11):1904–8. https://doi.org/10.1136/annrheumdis-2012-201746.
Wang B, Li Y, Wang H, Zhao J, Zhao Y, Liu Z, et al. FOXO3a is stabilized by USP18-mediated de-ISGylation and inhibits TGF-beta1-induced fibronectin expression. J Investig Med. 2020;68(3):786–91. https://doi.org/10.1136/jim-2019-001145.
Luciani A, Villella VR, Vasaturo A, Giardino I, Raia V, Pettoello-Mantovani M, et al. SUMOylation of tissue transglutaminase as link between oxidative stress and inflammation. J Immunol. 2009;183(4):2775–84. https://doi.org/10.4049/jimmunol.0900993.
Kusko RL, Brothers JF 2nd, Tedrow J, Pandit K, Huleihel L, Perdomo C, et al. Integrated genomics reveals convergent transcriptomic networks underlying chronic obstructive pulmonary disease and idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2016;194(8):948–60. https://doi.org/10.1164/rccm.201510-2026OC.
Nakajima F, Aratani S, Fujita H, Yagishita N, Ichinose S, Makita K, et al. Synoviolin inhibitor LS-102 reduces endoplasmic reticulum stress-induced collagen secretion in an in vitro model of stress-related interstitial pneumonia. Int J Mol Med. 2015;35(1):110–6. https://doi.org/10.3892/ijmm.2014.1984.
Nan L, Jacko AM, Tan J, Wang D, Zhao J, Kass DJ, et al. Ubiquitin carboxyl-terminal hydrolase-L5 promotes TGFbeta-1 signaling by de-ubiquitinating and stabilizing Smad2/Smad3 in pulmonary fibrosis. Sci Rep. 2016;6:33116. https://doi.org/10.1038/srep33116.
Pietkiewicz S, Sohn D, Piekorz RP, Grether-Beck S, Budach W, Sabapathy K, et al. Oppositional regulation of Noxa by JNK1 and JNK2 during apoptosis induced by proteasomal inhibitors. PLoS ONE. 2013;8(4):e61438. https://doi.org/10.1371/journal.pone.0061438.
Guo W, Shang F, Liu Q, Urim L, Zhang M, Taylor A. Ubiquitin-proteasome pathway function is required for lens cell proliferation and differentiation. Invest Ophthalmol Vis Sci. 2006;47(6):2569–75. https://doi.org/10.1167/iovs.05-0261.
Walter RFH, Sydow SR, Berg E, Kollmeier J, Christoph DC, Christoph S, et al. Bortezomib sensitivity is tissue dependent and high expression of the 20S proteasome precludes good response in malignant pleural mesothelioma. Cancer Manag Res. 2019;11:8711–20. https://doi.org/10.2147/CMAR.S194337.
Song K, Peng S, Sun Z, Li H, Yang R. Curcumin suppresses TGF-beta signaling by inhibition of TGIF degradation in scleroderma fibroblasts. Biochem Biophys Res Commun. 2011;411(4):821–5. https://doi.org/10.1016/j.bbrc.2011.07.044.
Pai CS, Khuat LT, Chen M, Murphy WJ, Abedi M. Therapeutic effects of a NEDD8-activating enzyme inhibitor, pevonedistat, on sclerodermatous graft-versus-host disease in mice. Biol Blood Marrow Transplant. 2017;23(1):30–7. https://doi.org/10.1016/j.bbmt.2016.10.022.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This work was supported by grants from the National Natural Science Foundation of China (NSFC; grant no. 81560502, 81960354), the Talent Project of Yunnan Province (2019HB024), the National Natural Science Foundation of Yunnan Province (grant no. 2019FE001-184), and the 100 Talents Program of Kunming Medical University (Lechun Lyu).
Conflicts of Interest
Wanlu Shen, Zhigang Zhang, Jiaqing Ma, Di Lu, and Lechun Lyu have no conflicts of interest that are directly relevant to the content of this article.
Ethics approval and Consent to participate
This article does not contain any studies with human participants or animals.
Consent for publication
All the authors listed have made full contributions to the project and can be included in the scope of authors.
Acknowledgements
The authors acknowledge the editors and reviewers for their positive and constructive comments and suggestions on our study.
Availability of data and materials
Not applicable.
Code availability
Not applicable.
Authors' contributions
Lechun Lyu and Di Lu contributed to the study conception and design. Wanlu Shen, Zhigang Zhang, Jiaqing Ma, Lechun Lyu and Di Lu prepared the main manuscript; All authors have read and approved the final article.
Rights and permissions
About this article
Cite this article
Shen, W., Zhang, Z., Ma, J. et al. The Ubiquitin Proteasome System and Skin Fibrosis. Mol Diagn Ther 25, 29–40 (2021). https://doi.org/10.1007/s40291-020-00509-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40291-020-00509-z