Abstract
Apoptosis was a term introduced in 1972 to distinguish a mode of cell death with characteristic morphology and apparently regulated, endogenously driven mechanisms. The effector processes responsible for apoptosis are now mostly well known, involving activation of caspases and Bcl2 family members in response to a wide variety of physiological and injury-induced signals. The factors that lead of the decision to activate apoptosis as opposed to adaptive responses to such signals (e.g. autophagy, cycle arrest, protein synthesis shutoff) are less well understood, but the intranuclear Promyelocytic Leukaemia Body (PML body) may create a local microenvironment in which the audit of DNA damage may occur, informed by the extent of the damage, the adequacy of its repair and other aspects of cell status.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Over the past two or three decades, a clear and seemingly comprehensive picture of the biology of apoptosis has emerged. Originally identified through its characteristic cytological morphology [1], this mode of death is now known to result from activation of a common mechanism relevant in both physiological and pathological circumstances [2, 3]. At the heart of this mechanism lie two families of proteins, the caspases and members of the Bcl2 extended family. Caspases are a unique and closely related set of proteases, so called because of the cysteine at their active site, and the tightly defined four-amino-acid motif (including aspartate at positions 1 and 4) at their target site. The Bcl2 family is so called because of the relationship of its members to the B-cell lymphoma oncogene whose discovery led eventually to the identification of most of the other family members, but at the molecular level this family is remarkably diverse.
Caspase Activation Underlies Most of the Phenotype of Apoptosis
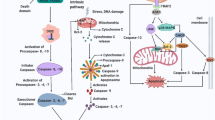
The caspases form a cascade in which initiator caspases are activated by lethal stimuli arising either at the cell membrane as a result of cytokine–receptor binding, or within the cell, in relation to internally determined signals, often generated in the micro-environment of particular organelles. Thus, caspases 8 and 10 are activated when specific extracellular ligands of the tumour necrosis factor family bind to their receptors (the extrinsic apoptosis pathway), whilst caspase 9 is activated at the mitochondrial membrane (the intrinsic pathway). These initiator caspases activate (by cleavage at their specific target sites) a set of effector caspases, notably caspases 3, 6 and 7, which then synchronously cleave proteins in many cell compartments. This cleavage event is responsible for most of the morphological changes by which apoptosis was originally identified. Thus, the violent blebbing of apoptotic cells is attributable to activation, by caspase cleavage, of the rho-kinase isoform ROCK-1 [4]. Caspase substrates also include cytoskeleton proteins [5] and the focal adhesion kinase (FAK) [6], whose cleavage accounts for the loss of substratum contact and the loss and rounding up of the dying cell. Some caspase substrates form part of complex pathways with several interacting members. One striking example is the cytoplasmic chaperone inhibitor of caspase-activated DNAse (ICAD), whose cleavage releases a nuclease from its anchor within the cytoplasm and permits its unfolding to reveal a nuclear localisation signal [7]. The unfolded active nuclease thus arrives within the nucleus where it is responsible for the cleavage and much of the condensation of nuclear DNA. Caspases may also be responsible for the release of nucleotides from apoptotic cells that serve as homing signals for the macrophages that ultimately engulf them [8]. Interestingly, the 4-amino-acid motifs that characterise caspase cleavage sites in scores if not hundreds of proteins [9] appear to be conserved between species as widely divergent as Drosophila, Xenopus and mammals [10, 11]. Thus, the activation of caspases is central to the synchronised molecular events that occur in apoptosis, although it must be said that it is still unclear whether some of these events are more “necessary and sufficient” than others in effecting the death of cells. A hint that this might be so is given by the observation that different members of the cascade appear to be preferentially selected in particular cell types and also show substrate specificity [12, 13].
The Bcl2 Family Members Define Thresholds for Apoptosis
The Bcl2 family members [2, 3, 14] are united through their possession of homologous domains responsible for protein–protein interactions amongst the family members. Bcl2 itself and its closest relatives (e.g. BclXL) possess four such domains, BH 1, 2 and 4, defining a hydrophobic groove within the molecule, and BH3, a short (8–12 amino acid) region that binds within that groove. These members of the family support cell survival, whilst the shared BH domains permit interaction with two powerful pro-death molecules, Bax and Bak, through the formation of heterodimers. Bax and Bak (which possess the BH1-3 domains but not BH4) can also form homo-oligomers and, in this configuration, can create a wide diameter pore through cell membranes [15]. This event has been extensively studied in the mitochondrial membrane, where such pores allow escape of critical molecules from the intermembranous space to the immediate peri-mitochondrial microenvironment. Amongst the escaping molecules are cytochrome c and dATP, which together activate caspase 9, held in this microenvironment by its association with a protein (with remarkable, seven-fold symmetry) called apaf-1. The concentrations of Bax or Bak relative to the Bcl2-BclXL or other pro-survival family members thus determine the probability that this dramatic rise in mitochondrial permeability will occur, activating the intrinsic pathway.
The remaining members of the Bcl2 family possess only the BH3 domain as their region of homology with the rest of this extended family. These proteins (bid, bad, bim, bmf and others) all promote death. Although their precise mode of action is still disputed, a likely explanation can be found in their high affinity of binding, via the BH3 domain, to the hydrophobic groove in Bcl2 and BclXL [14, 16]. A rise in cell concentration of BH3-only proteins will therefore create conditions in the immediate vicinity of the mitochondrial outer membrane that favour formation of bax/bak oligomer formation and the genesis of the high-permeability pores. This widely diverse family of “BH3-only” proteins appears to provide signals in response to a variety of injuries (Fig. 1). Thus, they act as sensors of “danger” or “stress” conditions.
Other Modes of Cell Death Exist
Apoptosis is widely observed in metazoans, but it is not the only route to cell death, even in the context of development. Moreover, even in organisms in which members of both the caspase and Bcl2 family are present constitutively, cells can sometimes undergo developmentally determined death despite experimental inhibition of caspases, showing that in these circumstances, the event of death must be determined by elements upstream of (or at least parallel to) the caspases themselves [17]. Interestingly, however, the structural changes in such dying cells differ from those of apoptosis and appear rather to represent loss of cellular volume homeostasis [18–20].
Despite the detail in which apoptosis is now understood, several major questions remain. Amongst these are two that are the subjects of the remainder of this short review. Both relate to apoptosis following cell injury. Under these circumstances, both in vivo and in vitro, it is usual to observe some cells entering apoptosis while their neighbours do not, despite being exposed to very similar lethal stimuli. The surviving cells often exhibit adaptive reactions that sustain cell life even in these unfavourable circumstances [21–23]. The questions therefore arise: first, what is the intracellular audit that determines which cells are selected for life, others for death? And second, what is the switch that determines that these adaptive changes are abandoned in favour of apoptosis? This article attempts to address both these questions, using cell damage by ionising radiation (IR) as the paradigm. The questions themselves, however, and hopefully their answers, are likely to be of general import.
The Search for an Intracellular Audit of DNA Damage
It is often assumed that apoptosis is initiated if the damage is “too severe to be repaired” or if the timescale for complete repair is “too long”. Although these statements seem probable, there is little evidence for the mechanism responsible. There is, however, excellent evidence that DNA repair is initiated swiftly after DNA damage by IR in cells destined to die by apoptosis a few hours later and is nearly complete before apoptosis is initiated. Thus, for example, comet assays show nearly complete restoration of supercoiled DNA within 1 hour of irradiation of bone-marrow-derived pre-B cells, whilst apoptosis is not evident until some 2 to 4 hours later, even under conditions in which more than 90% of the cells eventually die [24]. This suggests either that some lesion other than DNA breakage itself is involved in signalling apoptosis, or (following the basic design of other checkpoints) the defining event is the repair of the last persisting double-strand break (DSB), and failed completion of this, within some critical “window” in time, or perhaps in some other intracellular condition, spells death.
We argued that if the audit of cell injury following IR is based on DNA damage, its location is likely to be close to the damage itself, i.e. in an intranuclear site. This immediately contrasts with current paradigms, which, as outlined earlier, place the signals for apoptosis either at the cell membrane or in the vicinity of mitochondria. To be credible, a candidate audit mechanism for DNA damage would be expected to be responsive in a dose-dependent manner to the intranuclear DNA damage itself. Moreover, the audit apparatus would be expected to demonstrate a dose-related qualitative transition when responding to lethal as opposed to survivable levels of damage.
One candidate with these properties has been described—the Promyelocytic Leukaemia Nuclear Domain (PML-ND) or PML body. PML bodies are intranuclear particles consisting of a shell of around 0.2-μm diameter, constituted of polymerised sumoylated PML protein, together with a wide array of cargo proteins [25]. Most cells in culture have around 10 PML bodies per nucleus, although the precise number in any one population is influenced by cell type and position in the cell cycle. Notably, however, the number of PML bodies per nucleus rises dramatically following DNA damage, in human fibroblasts often by some 200% to 300% [26–29]. This increase peaks around 4 to 8 hours after DNA damage, reverting to near normal values by 12 to 24 hours if the IR dose is low (≤2.5 Gy), but remaining high if the IR dose is high (≥5 Gy). Interestingly, this transition corresponds to the transition between sublethal and lethal doses at least in terms of “reproductive death” (i.e. irreversible replication arrest). Another striking feature of PML bodies is their intranuclear location relative to the foci at which DNA damage and repair take place (IR-induced foci, or IRIFs). Initially following radiation, there is no particular spatial relationship between PML bodies and IRIFs, but within a few hours, most PML bodies are closely adjacent to IRIFs [26, 29] (Fig. 2). These features suggest, but do not prove, that the surface of PML- bodies is capable of identifying the status of damaged DNA as it undergoes repair.
The mechanism underlying this alteration in number of the bodies is not known for certain, but is considered to be the remit of protein modification rather than transcription or translational events, since it is not affected by blockade of protein synthesis by, for example, cycloheximide. Some data suggest that larger bodies split or bud to create increased numbers of smaller ones [30]. This might influence their capacity to permit interaction of their cargo proteins, or perhaps might transiently increase the reactive surface available for such interactions. However, this interpretation does not exclude other possibilities, such as PML-ND turnover, perhaps through differential sumoylation, in the vicinity of DSBs.
These observations are at least consistent with the hypothesis that PML bodies are a part of the nuclear response to DNA injury. Additional evidence comes from study of the nature of the cargo proteins, which include several involved in the response to injury, such as p53 (whose activation by acetylation and stabilisation by mdm2 appear to be facilitated by the presence of PML protein) [31–35], Blm (a helicase critical for completion of DSB repair) [36, 37] and Daxx (a modulator of apoptosis) [38]. Finally, and most significantly, tissues from animals genetically deficient in PML show attenuated apoptosis in response to a variety of lethal stimuli [39].
How might PML bodies become “aware” that the nucleus has sustained DNA damage?” The answer to this important question is not known, but a clue may be offered from consideration of the topographic molecular rearrangements that take place within the damaged nucleus. DSBs are swiftly reorganised by the DNA-dependent kinase ataxia telangiectasia mutated (ATM), which is phosphorylated close to the break site. Substrates of this kinase include many of the molecules that participate in DNA repair, such as the MRN complex, p53 BP1 and BRCA1 [40–42]. Detailed study of nuclear topology shows that these ATM substrates are activated in chromatin surrounding the break site, in association with the modified histone γH2AX [43]. Phosphorylation of this probably leads to the unwinding of chromatin that facilitates access of the repair molecules to the break site. A further class of repair-associated molecule is typified by the checkpoint-related kinases Chk-1 and Chk-2, which move freely throughout the damaged nucleus and act both as substrate for and a stimulus to some of the other phosphorylation events involved in repair. Interestingly, Chk-2-deficient cells show a grossly retarded PML response to DNA injury, suggesting that Chk-2 may be part of the signal that informs PML of the intranuclear presence of DNA DSBs.
The Switch From Adaptive Survival to Apoptosis
A still broader question asks what the relationship might be between the adaptive responses that support cell viability following injury and the homeostatic regulatory response that commits injured cells to death. The adaptive responses are heterogeneous, in that very different “stress” stimuli initiate stimulus-specific detection and immediate response mechanisms (Fig. 3). It is clear that, for example, quite distinctive intracellular networks are engaged in the reaction to endoplasmic reticulum stress (the unfolded protein response) [44–46], viral infection (including the interferon response) [47] and heat shock [48–50]. However, the ultimate cellular reactions appear to represent a common pathway, in which autophagy, protein synthesis shutoff and inhibition of DNA replication are conspicuous features. Although knowledge of the driving mechanism behind each of these fundamental adaptive responses to injury is still incomplete, a picture is emerging in which the PI3 kinase/Akt pathway plays a significant role in all three (Fig. 4). Interestingly, this pathway is also involved in the initiation of apoptosis and may include an intranuclear element that incorporates PML bodies in critical sites determining the outcome of Akt-driven phosphorylation [51].
A variety of different potentially lethal stimuli, each recognised by a specific set of transcription factors and signalling molecules, initiate the adaptive reactions of cycle arrest, autophagy and protein synthesis shutoff. For reasons that are still poorly understood, these reactions can be over-ruled by cell death, apparently by an endogenously controlled mechanism
One possible method to over-ride adaptive responses would be to recruit the innate immune system's killing power, through exposure of a “stress-dependent” or “danger” signal on the cell surface. One known example of this is the expression of the immunoglobulin-like molecules mic A and mic B on the surface of stressed cells [52]. These molecules (which are not expressed in this way in the absence of cellular stress) engage with NK cell receptors and permit activation of the latter's killing mechanism. It seems improbable, however, that a process as important as switching from survival to death should depend exclusively on a non-cell autonomous strategy such as NK cell activation.
Cell autonomous switches from adaptive responses to apoptosis in injured cells also exist. One example is the initiation of apoptosis following extremes of endoplasmic reticulum stress. Here, the transcription factor CHOP (for C/EBP homologous protein), a factor in the unfolded protein response, participates in the adaptation to protein overload, being transcriptionally activated in response to inducers of the unfolded protein response (ATF4, ATF6, XBP1) and also to ATF2, a molecular sensor for hypoxia and amino acid starvation. Over-expression of CHOP, however, initiates apoptosis [53, 54]. A mechanism of this type, in which signals emanating from within the stressed cell are both adaptive and pro-apoptotic, can be readily fitted into scenarios in which the permit to activate apoptosis is regulated through modification of the cell's apoptosis threshold—as described for BH3-only proteins or PML bodies above.
References
Kerr JF, Wyllie AH, Currie AR (1972) Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer 26:239–251
Danial NN, Korsmeyer SJ (2004) Cell death: critical control points. Cell 116:205–219
Hotchkiss RS, Strasser A, McDunn JE, Swanson PE (2009) Cell death. N Engl J Med 361:1570–1583
Coleman ML, Sahai EA, Yeo M, Bosch M, Dewar A, Olson MF (2001) Membrane blebbing during apoptosis results from caspase-mediated activation of ROCK-1. Nat Cell Biol 3:339–345
Ndozangue-Touriguine O, Hamelin J, Bréard J (2008) Cytoskeleton and apoptosis. Biochem Pharmacol 76:11–18
Wen L-P, Fahrni JA, Troie S, Guan J-L, Orth K, Rosen GD (1997) Cleavage of focal adhesion kinase by caspases during apoptosis. J Biol Chem 272:26056–26061
Nagata S (2000) Apoptotic DNA fragmentation. Exp Cell Res 256:12–18
Elliott MR, Chekeni FB, Trampont PC, Lazarowski ER, Kadi A, Walk SF, Park D, Woodson RI, Ostankovich M, Sharma P, Lysiak JJ, Harden TK, Leitinger N, Ravichandran KS (2009) Nucleotides released by apoptotic cells act as a find-me signal to promote phagocytic clearance. Nature 461:282–286
Fischer U, Jänicke RU, Schultze-Osthoff K (2003) Many cuts to ruin: a comprehensive update of caspase substrates. Cell Death Differ 10:76–100
Webb SJ, Nicholson D, Bubb VJ, Wyllie AH (1999) Caspase-mediated cleavage of APC results in an amino-terminal fragment with an intact armadillo repeat domain. FASEB J 13:339–346
Aravind L, Dixit VM, Koonin EV (1999) The domains of death: evolution of the apoptosis machinery. TIBS 24:47–53
Walsh JG, Cullen SP, Sheridan C, Lüthl AK, Gerner C, Martin SJ (2008) Executioner caspase-3 and caspase-7 are functionally distinct proteases. Proc Natl Acad Sci USA 105:12815–12819
Slee EA, Adrain C, Martin SJ (2001) Executioner caspase-3, -6, and -7 perform distinct, non-redundant roles during the demolition phase of apoptosis. J Biol Chem 276:7320–7326
Adams JM, Cory S (2007) Bcl-2-regulated apoptosis: mechanisms and therapeutic potential. Curr Opin Immunol 19:488–496
Kuwana T, Mackey MR, Perkins G, Ellisman MH, Latterich M, Schneiter R, Green DR, Newmeyer DD (2002) Bid, bax and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell 111:331–342
Willis SN, Fletcher JI, Kaufmann T, van Delft MF, Chen L, Czabotav PE, Ierino H, Lee EF, Fairlie WD, Bouillet P, Strasser A, Kluk RM, Adams JM, Huang DC (2007) Apoptosis initiated when BH3 ligands engage multiple Bcl-2 homologs, not Bax or Bak. Science 315:856–859
Chautan M, Chazal G, Cecconi F, Gruss P, Golstein P (1999) Interdigital cell death can occur through a necrotic and caspase-independent pathway. Curr Biol 9:967–970
Vandenabele P, Vanden Berghe, Festjens N (2006) Caspase inhibitors promote alternative cell death pathways. Sci STKE 2006 pe44
Kroemer G et al (2009) Classification of Cell Death: recommendations of the nomenclature committee on cell death 2009. Cell Death Differ 16:3–11
Wyllie AH, Golstein P (2001) More than one way to go. Proc Natl Acad Sci USA 98:11–13
Liebermann DA, Hoffman B (2008) Gadd 45 in stress signalling. J Molec Signalling 3:15–23
Cecconi F, Levine B (2008) The role of autophagy in mammalian development: cell makeover rather than cell death. Dev Cell 15:344–357
Siafakis AR, Richardson DR (2009) Growth Arrest and DNA damage—45 alpha (GADD 45 alpha). Int J Biochem Cell Biol 41:986–989
Griffiths SD, Clarke AR, Healy LE, Ross G, Ford AM, Hooper ML, Wyllie AH, Greaves M (1997) Absence of p53 permits propagation of mutant cells following genotoxic damage. Oncogene 14:523–531
Bernardi R, Pandolfi PP (2007) Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol 8:1006–1016
Varadaraj A, Dovey CL, Laredj L, Ferguson B, Alexander CE, Lubben N, Wyllie AH, Rich T (2007) Evidence for the receipt of DNA damage stimuli by PML nuclear domains. J Pathol 211:471–480
Salomoni P, Ferguson BJ, Wyllie AH, Rich T (2008) New insights into the role of PML in tumour suppression. Cell Res 18:622–640
Dellaire G, Ching RW, Ahmed K, Jalali F, Tse KC, Bristow RG, Bazett-Jones DP (2006) Promyelocytic nuclear bodies behave as DNA damage sensors whose response to DNA double-strand breaks is regulated by NBS-1 and the kinases ATM, Chk2 and ATR. J Cell Biol 175:55–66
Dellaire G, Kepkay R, Bazett-Jones DP (2009) High resolution imaging of changes in the structure and spatial organisation of chromatin, gamma-H2AX and the MRN complex within etoposide-induced DNA repair foci. Cell Cycle 8:3750–3769
Dellaire G, Ching RW, Dehghani H, Ren Y, Bazett-Jones DP (2006) The number of PML nuclear bodies increases in early S phase by a fission mechanism. J Cell Sci 119:1026–1033
Pearson M, Carbone R, Sebastiani C, Cioce M, Fagioli M, Saito S, Higashimoto Y, Appella E, Minucci S, Pandolfi PP, Pelicci PG (2000) PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature 406:207–210
Gu OA, Salamoni P, Luo J, Shih A, Zhong S, Gu W, Pandolfi PP (2000) The function of PML in p53-dependent apoptosis. Nat Cell Biol 2:730–736
Fogal V, Gostissa M, Sandy P, Zacchi P, Sternsdorf T, Jensen K, Pandolfi PP, Will H, Schneider C, Del Sal F (2000) Regulation of p53 activity in nuclear bodies by a specific PML isoform. EMBO J 19:6185–6195
Bernardi R, Scaglioni PP, Bergmann S, Horn HF, Vousden KH, Pandolfi PP (2004) PML regulates p53 stability by sequestering Mdm2 to the nucleus. Nat Cell Biol 6:665–672
de Stanchina E, Querido E, Narita M, Davuluri RV, Pandolfi PP, Ferbeyre G, Lowe SW (2004) PML is a direct p53 target that modulates p53 effector functions. Mol Cell 13:523–535
Bischof O, Kim SH, Irving J, Beresten S, Ellis NA, Campisi J (2001) Regulation and localization of the Bloom syndrome protein in response to DNA damage. J Cell Biol 153:367–380
Wang XW, Tseng A, Ellis NA, Spillare EA, Linke SP, Robles AI, Seker H, Yang Q, Hu P, Beresten S, Bemmels NA, Garfield S, Harvis CC (2001) Functional interaction of p53 and BLM DNA helicase in apoptosis. J Biol Chem 276:32948–32955
Ishov AM, Sothikov AG, Negorev D, Vladimirova OV, Neff N, Kamitani T, Yeh ET, Strauss JF, Maul GG (1999) PML is critical for ND10 formation and recruits the PML-intereacting protein daxx to this nuclear structure when modified by SUMO-1. J Cell Biol 147:221–234
Bernardi R, Pandolfi PP (2003) Role of PML and the PML nuclear body in the control of cell death. Oncogene 22:9048–9057
Khanna KK, Lavin MF, Jackson SP, Mulhern TD (2001) ATM, a central controller of cellular responses to DNA damage. Cell Death Differ 8:1052–1055
Stucki M, Jackson SP (2006) Gamma H2AX and MDC1: anchoring the DNA-damage-response machinery to broken chromosomes. DNA Repair (Amst) 5:534–543
Riches LG, Lynch AM, Goodesham NJ (2008) Early events in the mammalian response to DNA double-strand breaks. Mutagenesis 23:331–334
Bekker-Jensen S, Lukas C, Kitagawa R, Melander F, Kastan M, Bartek J, Lukas J (2006) Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J Cell Biol 173:195–206
Kim I, Xu W, Reed JC (2008) Cell death and endoplasmic reticulum stress, disease relevance and therapeutic opportunities. Nat Rev Drug Discov 7:1013–1030
Rasheva VI, Domingos PM (2009) Cellular responses to endoplasmic reticulum stress and apoptosis. Apoptosis 14:996–1007
Kohno K (2010) Stress-sensing mechanisms in the unfolded protein response: similarities and differences between yeast and mammals. J Biochem 147:27–33
Tacheuchi O, Akira S (2009) Innate immunity to virus infection. Immunol Rev 227:75–86
Liu X-D, Li PCC, Santoro N, Thiele DJ (1997) Conservation of a stress response: human heat shock transcription factors functionally substitute for yeast HSF. EMBO J 16:6466–6477
Dice JF (2007) Chaperone-mediated autophagy. Autophagy 3:295–299
Ravikumar B et al (2009) Mammalian macroautophagy at a glance. J Cell Sci 122:1707–1711
Trotman LC, Alimonti A, Scaglioni PP, Koutcher JA, Gordon-Cardo C, Pandolfi PP (2006) Identification of a tumour suppressor network opposing nuclear Akt function. Nature 441:523–527
Steinle A, Li P, Morris DL, Groh V, Lanier LL, Strong RK, Spies T (2001) Interactions of human NKG2D with its ligands MICA and MIC B and homologues of the mouse RAE-3 family. Immunogenetics 53:279–287
Marciniak SJ, Yun CY, Oyadomari S, Novoa I, Zhang Y, Hungreis R, Nagata K, Harding HP, Ron D (2004) CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev 18:3066–3077
Song B, Scheuner D, Ron D, Pennathur S, Kaufman RJ (2008) CHOP deletion reduces oxidative stress, improves beta cell function and promotes cell survival in multiple mouse models of diabetes. J Clin Invest 118:3378–3389
Acknowledgments
AHW’s work was supported by the UK Biotechnology and Biological Sciences Research Council (BBSRC).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Wyllie, A.H. “Where, O Death, Is Thy Sting?” A Brief Review of Apoptosis Biology. Mol Neurobiol 42, 4–9 (2010). https://doi.org/10.1007/s12035-010-8125-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-010-8125-5