Abstract
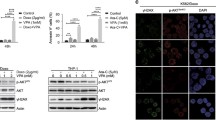
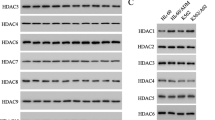
Cytarabine-resistant acute myeloid leukemia (AML) is a common phenomenon, necessitating the search for new chemotherapeutics. WEE1 participates in cell cycle checkpoint signaling and inhibitors targeting WEE1 (WEE1i) constitute a potential novel strategy for AML treatment. HDAC (histone deacetylase) inhibitors have been shown to enhance the anti-tumor effects of WEE1i but molecular mechanisms of HDAC remain poorly characterized. In this study, the WEE1 inhibitor PD0166285 showed a relatively good anti-leukemia effect. Notably, PD0166285 can arise the expression of HDAC11 which was negatively correlated with survival of AML patients. Moreover, HDAC11 can reduced the anti-tumor effect of PD0166285 through an effect on p53 stability and the changes in phosphorylation levels of MAPK pathways. Overall, the cell cycle inhibitor, PD0166285, is a potential chemotherapeutic drug for AML. These fundings contribute to a functional understanding of HDAC11 in AML.






Similar content being viewed by others
Data availability
All data from UCSC Xena database are publicly available. The main data from this manuscript are available from the corresponding author on reasonable request.
References
Aleem E, Arceci RJ. Targeting cell cycle regulators in hematologic malignancies. Front Cell Dev Biol. 2015;3:16.
Lin Y, Kang T, Zhou BP. Doxorubicin enhances Snail/LSD1-mediated PTEN suppression in a PARP1-dependent manner. Cell Cycle. 2014;13(11):1708–16.
Niida H, Katsuno Y, Banerjee B, Hande MP, Nakanishi M. Specific role of Chk1 phosphorylations in cell survival and checkpoint activation. Mol Cell Biol. 2007;27(7):2572–81.
Garcia TB, Snedeker JC, Baturin D, et al. A small-molecule inhibitor of WEE1, AZD1775, synergizes with olaparib by impairing homologous recombination and enhancing DNA damage and apoptosis in acute leukemia. Mol Cancer Ther. 2017;16(10):2058–68.
Zhou L, Zhang Y, Chen S, et al. A regimen combining the Wee1 inhibitor AZD1775 with HDAC inhibitors targets human acute myeloid leukemia cells harboring various genetic mutations. Leukemia. 2015;29(4):807–18.
Pai JT, Hsu CY, Hua KT, et al. NBM-T-BBX-OS01, semisynthesized from osthole, induced G1 growth arrest through HDAC6 inhibition in lung cancer cells. Molecules. 2015;20(5):8000–19.
Long J, Fang WY, Chang L, et al. Targeting HDAC3, a new partner protein of AKT in the reversal of chemoresistance in acute myeloid leukemia via DNA damage response. Leukemia. 2017;31(12):2761–70.
Long J, Jia MY, Fang WY, et al. FLT3 inhibition upregulates HDAC8 via FOXO to inactivate p53 and promote maintenance of FLT3-ITD+ acute myeloid leukemia. Blood. 2020;135(17):1472–83.
Johansson AC, Ansell A, Jerhammar F, et al. Cancer-associated fibroblasts induce matrix metalloproteinase-mediated cetuximab resistance in head and neck squamous cell carcinoma cells. Mol Cancer Res. 2012;10(9):1158–68.
Huang F, Sun J, Chen W, et al. HDAC4 inhibition disrupts TET2 function in high-risk MDS and AML. Aging (Albany NY). 2020;12(17):16759–74.
Sharma V, Wright KL, Epling-Burnette PK, Reuther GW. Metabolic vulnerabilities and epigenetic dysregulation in myeloproliferative neoplasms. Front Immunol. 2020;11:604142.
Bi L, Ren Y, Feng M, et al. HDAC11 regulates glycolysis through the LKB1/AMPK signaling pathway to maintain hepatocellular carcinoma stemness. Cancer Res. 2021;81(8):2015–28.
Yue L, Sharma V, Horvat NP, et al. HDAC11 deficiency disrupts oncogene-induced hematopoiesis in myeloproliferative neoplasms. Blood. 2020;135(3):191–207.
Bora-Singhal N, Mohankumar D, Saha B, et al. Novel HDAC11 inhibitors suppress lung adenocarcinoma stem cell self-renewal and overcome drug resistance by suppressing Sox2. Sci Rep. 2020;10(1):4722.
Xiao C, Wang Y, Zheng M, et al. RBBP6 increases radioresistance and serves as a therapeutic target for preoperative radiotherapy in colorectal cancer. Cancer Sci. 2018;109(4):1075–87.
Chao CC. Mechanisms of p53 degradation. Clin Chim Acta. 2015;438:139–47.
Saygin C, Carraway HE. Emerging therapies for acute myeloid leukemia. J Hematol Oncol. 2017;10(1):93.
Winer ES, Stone RM. Novel therapy in acute myeloid leukemia (AML): moving toward targeted approaches. Ther Adv Hematol. 2019;10:2040620719860645.
Lockhead S, Moskaleva A, Kamenz J, et al. The apparent requirement for protein synthesis during G2 phase is due to checkpoint activation. Cell Rep. 2020;32(2):107901.
Tsai TY, Theriot JA, Ferrell JE Jr. Changes in oscillatory dynamics in the cell cycle of early Xenopus laevis embryos. PLoS Biol. 2014;12(2):e1001788.
Mir SE, De Witt Hamer PC, Krawczyk PM, et al. In silico analysis of kinase expression identifies WEE1 as a gatekeeper against mitotic catastrophe in glioblastoma. Cancer Cell. 2010;18(3):244–57.
Gao L, Cueto MA, Asselbergs F, Atadja P. Cloning and functional characterization of HDAC11, a novel member of the human histone deacetylase family. J Biol Chem. 2002;277(28):25748–55.
Thangapandian S, John S, Lee Y, Arulalapperumal V, Lee KW. Molecular modeling study on tunnel behavior in different histone deacetylase isoforms. PLoS ONE. 2012;7(11):e49327.
Deubzer HE, Schier MC, Oehme I, et al. HDAC11 is a novel drug target in carcinomas. Int J Cancer. 2013;132(9):2200–8.
Bradbury CA, Khanim FL, Hayden R, et al. Histone deacetylases in acute myeloid leukaemia show a distinctive pattern of expression that changes selectively in response to deacetylase inhibitors. Leukemia. 2005;19(10):1751–9.
Yu Z, Wang R, Chen F, Wang J, Huang X. Five novel oncogenic signatures could be utilized as AFP-related diagnostic biomarkers for hepatocellular carcinoma based on next-generation sequencing. Dig Dis Sci. 2018;63(4):945–57.
Gong D, Zeng Z, Yi F, Wu J. Inhibition of histone deacetylase 11 promotes human liver cancer cell apoptosis. Am J Transl Res. 2019;11(2):983–90.
Wang W, Fu L, Li S, Xu Z, Li X. Histone deacetylase 11 suppresses p53 expression in pituitary tumor cells. Cell Biol Int. 2017;41(12):1290–5.
Li J, Wang Y, Sun Y, Lawrence TS. Wild-type TP53 inhibits G(2)-phase checkpoint abrogation and radiosensitization induced by PD0166285, a WEE1 kinase inhibitor. Radiat Res. 2002;157(3):322–30.
Wang Y, Li J, Booher RN, et al. Radiosensitization of p53 mutant cells by PD0166285, a novel G(2) checkpoint abrogator. Cancer Res. 2001;61(22):8211–7.
Devine T, Dai MS. Targeting the ubiquitin-mediated proteasome degradation of p53 for cancer therapy. Curr Pharm Des. 2013;19(18):3248–62.
Zhou R, Wu J, Tang X, et al. Histone deacetylase inhibitor AR-42 inhibits breast cancer cell growth and demonstrates a synergistic effect in combination with 5-FU. Oncol Lett. 2018;16(2):1967–74.
Ito A, Kawaguchi Y, Lai CH, et al. MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J. 2002;21(22):6236–45.
Li M, Luo J, Brooks CL, Gu W. Acetylation of p53 inhibits its ubiquitination by Mdm2. J Biol Chem. 2002;277(52):50607–11.
Liu Z, Wang Y, Gao T, et al. CPLM: a database of protein lysine modifications. Nucleic Acids Res. 2014;42:D531-36.
Wang G, Li S, Gilbert J, et al. Crucial roles for SIRT2 and AMPA receptor acetylation in synaptic plasticity and memory. Cell Rep. 2017;20(6):1335–47.
Caron C, Boyault C, Khochbin S. Regulatory cross-talk between lysine acetylation and ubiquitination: role in the control of protein stability. BioEssays. 2005;27(4):408–15.
Buglio D, Khaskhely NM, Voo KS, Martinez-Valdez H, Liu YJ, Younes A. HDAC11 plays an essential role in regulating OX40 ligand expression in Hodgkin lymphoma. Blood. 2011;117(10):2910–7.
Thole TM, Lodrini M, Fabian J, et al. Neuroblastoma cells depend on HDAC11 for mitotic cell cycle progression and survival. Cell Death Dis. 2017;8(3):e2635.
Acknowledgements
We also thank the UCSC Xena database for the contribution in our study.
Funding
This research was supported by grants from the National Natural Science Foundation of China (Grant Number 81772280), Key Technology Innovation Special of Key Industries of the Chongqing Science and Technology Bureau (Grant Number csct2022ycjh-bgzxm0034), and Chongqing Graduate Research Innovation Project Funding (CYS21229).
Author information
Authors and Affiliations
Contributions
Conceptualization: ZZ and LZ; investigation: ZZ, XC, and WD; resources: ZZ and PW; data curation: ZZ, XS, and SC; writing—original draft: ZZ; writing—review & editing: ZZ and YL; supervision: BL; project administration: BL.
Corresponding author
Ethics declarations
Conflict of interest
The author declares no conflicts of this manuscript.
Research involving human and animal rights
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent to participate
N/A.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhou, Z., Zhong, L., Chu, X. et al. HDAC11 mediates the ubiquitin-dependent degradation of p53 and inhibits the anti-leukemia effect of PD0166285. Med Oncol 40, 325 (2023). https://doi.org/10.1007/s12032-023-02196-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-023-02196-2