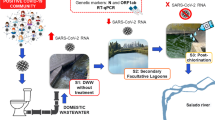
Abstract
The presence of SARS-CoV-2 RNA has been extensively reported at the influent of wastewater treatment plants (WWTPs) worldwide, and its monitoring has been proposed as a potential surveillance tool to early alert of epidemic outbreaks. However, the fate of the SARS-CoV-2 RNA in the treatment process of WWTP has not been widely studied yet; therefore, in this study, we aimed to evaluate the efficiency of treatment processes in reducing SARS-CoV-2 RNA levels in wastewater. The treatment process of three WWTPs of the Parisian area in France was monitored on six different weeks over a period of 2 months (from April 14 to June 9, 2021). SARS-CoV-2 RNA copies were detected using digital polymerase chain reaction (dPCR). Investigation on the presence of variants of concern (Del69-70, E484K, and L452R) was also performed. Additionally, SARS-CoV-2 RNA loads in the WWTPs influents were expressed as the viral concentration in per population equivalent (PE) and showed a good correlation with French public health indicators (incidence rate). SARS-CoV-2 RNA loads were notably reduced along the water treatment lines of the three WWTPs studied (2.5–3.4 log reduction). Finally, very low SARS-CoV-2 RNA loads were detected in effluents (non-detected in over half of the samples) which indicated that the potential risk of the release of wastewater effluents to the environment is probably insignificant, in the case of WWTPs enabling an efficient biological removal of nitrogen.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Surveillance of different markers in WWTPs influents has enabled in the past to characterize emerging chemicals and illicit drug use patterns and food consumption patterns (nitrogen and phosphorus) (Bressy et al. 2016; Gasperi et al. 2008; Rocher and Azimi 2016). Recently, the surveillance of SARS-CoV-2 has contributed to the understanding on the disease spread within the communities (Ahmed et al. 2020a; Kumar et al. 2021a; Medema et al. 2020; Randazzo et al. 2020; Wurtzer et al. 2020). In August 2020, the World Health Organization confirmed the presence of SARS-CoV-2 viral RNA in wastewater influent and in sewage sludge from several cities around the world (Milan, Paris, Murcia, Brisbane, Connecticut, and Massachusetts, among others) (WHO 2020). Monitoring the behavior of SARS-CoV-2 RNA within sewer systems rapidly appeared to be an interesting tool that provides precious information on the health of entire communities, however, the efficiency of WWTP treatment processes and the potential health risks associated with the release of wastewater effluents containing SARS-CoV-2 into the environment remain to be verified (Giacobbo et al. 2021). As the COVID-19 epidemy continues to spread all over the world, new variants of the SARS-CoV-2 virus are being detected. These variants might be more transmissible or capable of evading immune response, or their mutations might suppress diagnostic detection (Ascoli 2021; Singh et al. 2021; Wurtzer et al. 2020).
Digital PCR (dPCR) was rapidly pointed out as a good candidate regarding its sensitivity and quantification accuracy for SARS-CoV-2 monitoring (Staley et al. 2018; Rački et al. 2014; Hart and Halden 2020). Cao et al. have indeed shown in 2015 that dPCR exhibited higher precision and reproducibility than qPCR regarding quantification of human-associated fecal indicators in water (Cao et al. 2015). Besides quantification method, several authors have shown that virus concentration methods were also a critical aspect for an accurate and sensible quantification of SARS-CoV-2 in raw wastewater (Ahmed et al. 2020b; Lu et al. 2020; Jafferali et al. 2021). A European patent from the 31st of December 2020 under the application number EP20306715.2 was thus developed by researchers of Ingénierie et Analyse en Génétique Environnementale (I.A.G.E) to have a reliable process for virus quantification in liquid matrices (comprising sampling, extraction, and quantification steps). This method includes various optimization and quality control steps, which are crucial for generating reliable public health information as shown by Ahmed et al. (2020c) and Berestycki et al. (2021). Furthermore, as variants of concern SARS-CoV-2 have emerged more recently (Del69-70, E484K, and L452R, among others), dedicated tools to target and monitor the variants were also developed.
The present work intended to (i) provide an insight of the evolution of the main variants present in the raw waters coming from three different urban catchments and (ii) evaluate the presence of SARS-CoV-2 RNA using the new method of dPCR through the treatment process in three majors Parisian WWTPs.
Materials and methods
Parisian WWTPs monitored in this study
The three studied WWTPs are located upstream and downstream of the Parisian conurbation, and their flows vary between 50,000 and 600,000 m3 per day (Fig. 1). All of them are operated by the Greater Paris Sanitation Authority (SIAAP) in charge of collecting, transporting, and treating wastewater produced by close to 9 millions of human population. Different technologies are used for the treatment of water and sludge. The Seine Gresillons (SEG) plant uses the biofiltration process for water treatment, the Seine Valenton plant (SEV) uses activated sludge treatment for water, and the Seine Morée plant (SEM) uses membrane filtration for water treatment. These WWTPs are designed to efficiently reach the European standards (« Directive 91/271/EEC» 1991; « Directive 2000/60/EC» 2000).
These three plants are fed by raw water coming from urban catchments presenting contrasted characteristics and differ mainly in their design capacity and treatment processes, as briefly described in Table 1.
The catchment area of SEG (located in the northwest area) is characterized by a very high urbanization level and by contrast, SEV and SEM (located in the southeast and east area) have a moderate urbanization level.
Data treatment
Log removal values were calculated as follows:
Sewage sampling and analytical procedures
Samples of raw (influent), settled, and treated (effluent) wastewaters were collected using auto-sampler devices, with a flow-paced sampling program during dry-weather conditions, from 7 to 7 am the next day. A first campaign took place from April 14 to April 28, 2021, and a second campaign from May 26 to June 9, 2021, corresponding to contrasting SARS-CoV-2 incidence levels (French National Health Agency).
-
Sample treatment
To detect SARS-CoV-2 RNA in wastewater samples, we performed a diagnostic method recently developed to detect very low concentrations of SARS-CoV-2 in wastewater samples. This method combines an ultrafiltration and an optimized extraction process (This method has been submitted by IAGE to the European Patent Office on the 31st of December 2020 under the application number EP20306715.2) with a DNA quantification based on dPCR. For each wastewater sample of 1L, one RNA extraction was done on 30 mL concentrate. The method parameters were optimized using different types of wastewater (from WWTP with nominal capacity from 10 000PE to 400 000PE) spiked with attenuated SARS-CoV-2.
-
Sample analysis
The RT-dPCR (RetroTranscriptase-digital PCR) reaction was performed following the manufacturer’s instructions (QIAGEN, Germany) using the QIAcuity Eight Platform System, 5-plex (Cat. No. 911052), the QIAcuity One-Step Viral RT-PCR Kit (Cat. No. 1123145), and QIAcuity Nanoplate 26 K 24-well (Cat. No. 250001). A total of 26,000 RT-dPCR reactions by RNA extractions from 30 mL of concentrated wastewater were performed.
The RT-dPCR reaction mixture for SARS-CoV-2 variant strain detection was prepared in a pre-plate as follows: depending on nanoplate type. For Nanoplate 26 K reactions, 10 μl of 4 × One-Step Viral RT-PCR Master Mix, 0.4 μl of 100 × Multiplex Reverse Transcription Mix, 5 μl of the primers/probes mix from the PENTA-CoV wastewater Kit (00,283), 4 μl of RNA extract, and RNase-free water were combined to reach a final reaction volume of 40 μl. The mixture was prepared in a pre-plate and then transferred into the 24-well 26 K Nanoplate. The latter was then loaded to the QIAcuity 8 instrument, which is a fully automated system. The workflow included (i) priming and rolling step in order to generate and isolate the chamber partitions; (ii) the amplification step under the following cycling protocol: 50 °C for 40 min for reverse transcription, 95 °C for 2 min for enzyme activation, 95 °C for 5 s for denaturation, and 58 °C for 60 s for annealing/extension for 40 cycles; and (iii) the imaging step was done by reading.
To screen for important SARS-CoV-2 variants, particular molecular signatures were developed using a 5-plex assay that takes full advantage of the five detection channels available on the QIAcuity One 5plex, QIAcuity Four, and QIAcuity Eight instruments. The 5-plex assay uses one probe to detect SARS-CoV-2 wild-type N1 region NC_045512v2; a second probe to detect the Del H69-V70 mutations associated with the so-called Alpha variant: 20I/501Y.V1 (B.1.1.7); a third probe to detect the L452R mutation associated mainly with global variants: VOC (B.1.617.2), Delta Variant; a fourth probe to detect the E484K mutation mainly associated with the Beta and Gamma variants: 20H/501Y.V2 (B.1.351), 20 J/501Y.V3 (P.1); and a fifth probe targeting Pepper mild mottle virus (PMMoV) (NC_003630) that serves as a human density control (see Table S1 in Supplementary information). Limit of quantification (LQ) was 550 GU/L.
Physicochemical quality parameters were collected from the WWTPs through regulatory monitoring in raw and treated wastewater. These parameters were measured on a daily basis on 24-h composite samples collected with automated samplers. Analyses were performed by SIAAP central laboratory according to the following norms: NF EN 872 for suspended solids, SS, NF EN ISO 5815–1 for biochemical oxygen demand and BOD5, and NF EN ISO 12260 for total nitrogen, TN.
Results and discussion
SARS-CoV-2 RNA concentration dynamic in raw waters
Table 2 summarizes the SARS-CoV-2 RNA loads obtained using RT-dPCR technique in the three studied sewage Parisian WWTPs.
During the first sampling campaign period (from 14 to 28 April 2021), high concentrations of SARS-CoV-2 RNA were detected in the influents of Parisian WWTPs, with average values of 236 GU/mL for SEV, 273 GU/mL for SEG, and 481 GU/mL for SEM. Lower concentrations were obtained during the second sampling campaign period (from 26 May to 9 June 2021) with average concentrations of 41 GU/mL for SEV, 25 GU/mL for SEG, and 50 GU/mL for SEM. The decrease of the SARS-CoV-2 concentration from April to June 2021 is in good agreement with the reports of the French National Health Agency showing a decrease of the incidence rate of the epidemic available as open access data: https://www.data.gouv.fr/fr/datasets/synthese-des-indicateurs-de-suivi-de-lepidemie-covid-19/.
The normalized SARS-CoV-2 RNA concentration (per 100 000 PE) and the incidence rate (per 100 000 hab.) for the departments corresponding to the catchment area of each WWTP are also presented in Table 2. The incidence rate data of departments 75 (Paris), 92 (Hauts-de-France), 93 (Seine- Saint Denis), and 94 (Val-de-Marne) were collected from the open data portal of the French National Health Agency. As broadly discussed in other studies, the correlation between the SARS-CoV-2 RNA concentration in raw wastewater and the incidence rate (r2 = 0.61, Fig. S1) confirms that SARS-CoV-2 RNA monitoring in wastewater is a good candidate indicator of the epidemic spread, as recently shown by Ahmed et al. (2020a), Balboa et al. (2020), Medema et al. (2020), and Wurtzer et al. (2020).
Besides the quantification of SARS-CoV-2, we determined the presence of variants of concern within the population: Alpha (Del69-70), Beta-Gamma (E484), and Delta (L452R). The proportion of Alpha and Beta-Gamma variants respectively associated with Del69-70 and E484 mutations detected during the first sampling period is in good agreement with the open data published weekly by the French National Health Agency. However, the Delta variant L452R was not a variant of concern during the sampling period; therefore, no data related to it were published at that time. It can be noted that the L452R variant was already present on influent samples from April 28, 2021, in the Parisian region; moreover, similar proportions of L452R variant (23%) were reached among sequenced patients swabs only from June 20 to 27, 2021, in the Parisian region. These results indicate that SARS-CoV-2 RNA isolated from WWTP influents is also a reliable tool to detect the introduction of variants of concern in the local population weeks before they appear at significant levels in either clinical or screening swab samples (Bar-Or et al. 2021; Buenestado-Serrano et al. 2021; Carcereny et al. 2022; Heijnen et al. 2021; Li et al. 2021; Peterson et al. 2022; Wurtzer et al. 2022; Yaniv et al. 2021a, b).
Efficiency of treatment process from WWTPs
SARS-CoV-2 RNA average removal efficiency of the global treatment process as well as of the settling and biological treatment steps between April 14 and April 28 are shown in Fig. 2a. The efficiency of WWTPs on reducing the values of main physicochemical parameters is presented in Fig. 2b.
The three WWTP allows an efficient removal of particles, organic matter, and nitrogen. Indeed, the treatment water processes enables the removal of 97–99% of suspended solids, 96–100% of BOD5, and 73–85% of total nitrogen, as shown by Fig. 2b. In these operating conditions, the average reduction of SARS-CoV-2 RNA was of 1.60–2.06 log reduction as it can be seen in Fig. 2a. Regarding the settling step, low to moderate reductions of SARS-CoV-2 RNA levels were observed (0.01–0.26 log reduction), whereas for the biological treatment step, higher removals were obtained (1.6–2.3 log reduction). Low concentrations (< 50 GU/ml) of SARS-CoV-2 RNA were detected in outlets of the studied WWTPs, in all cases except on April 28 in SEV (Table 2).
Recent studies on the reduction of SARS-CoV-2 RNA in WWTPs (Hong et al. 2021; Kumar et al. 2021b; Serra-Compte et al. 2021) have reported removal efficiencies (0.5–1.98log) slightly lower than the present study. However, no fair and deeper comparisons can be established since treatment processes and influent quality differ notably. Some authors (Balboa et al. 2020; Kitamura et al. 2021; Kocacemi et al. 2020; Kumar et al. 2021a; Li et al. 2021) have detected high concentrations of SARS-CoV-2 RNA in wastewater sludge and hypothesized that viral material is mainly accumulated the solid fraction which implies that sludge treatment could efficiently removes SARS-CoV-2 from wastewater.
Conclusions
The presence of SARS-CoV-2 RNA in raw wastewater (influent) and settled and treated water (effluent) was quantified using RT-dPCR technique. SARS-CoV-2 RNA loads on influent showed good correlation with the incidence rate of COVID-19 in the Parisian area. The presence of variant L452R (Delta) was detected in samples from the present study from April 18, a few weeks prior to its inclusion as a variant of concern by the French National Health Agency, which confirms the interest of using sewage analysis as a complementary approach to early detection of epidemics outbreaks.
Finally, the three standard sewage treatment processes (activated sludge, biofiltration, or membrane bioreactor) in the Parisian area operated by SIAAP, with a complete treatment of carbon and nitrogen, are very efficient in eliminating SARS-CoV-2 RNA, with average reductions of 1.60–2.06 log.
By way of perspective, a comparison with other viruses on previous study on the Parisian WWTPs shows that the overall efficiency of treatment process is equivalent for F-specific RNA Bacteriophages (2.7–3.4 log reduction) (Mailler et al. 2021; Rocher and Azimi 2016). Results on the comparable removal of SARS-CoV-2 RNA and F-specific RNA Bacteriophages were recently reported (Montier et al. 2021; Serra-Compte et al. 2021). Further investigations should be performed to validate the use of RNA-F bacteriophages as indicators of SARS-CoV-2 removal along WWTPs.
Data availability
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
References
Ahmed W, Angel N, Edson J, Bibby K, Bivins A, O’Brien JW, Choi PM, Kitajima M, Simpson SL, Li J, Tscharke B, Verhagen R, Smith WJM, Zaugg J, Dierens L, Hugenholtz P, Thomas KV, Mueller JF (2020a) First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci Total Environ 728:138764. https://doi.org/10.1016/j.scitotenv.2020.138764
Ahmed W, Bertsch PM, Bivins A, Bibby K, Farkas K, Gathercole A, Haramoto E, Gyawali P, Korajkic A, McMinn BR, Mueller JF, Simpson SL, Smith WJM, Symonds EM, Thomas KV, Verhagen R, Kitajima M (2020b) Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci Total Environ 739:139960. https://doi.org/10.1016/j.scitotenv.2020.139960
Ahmed W, Bivins A, Bertsch PM, Bibby K, Choi PM, Farkas K, Gyawali P, Hamilton KA, Haramoto E, Kitajima M, Simpson SL, Tandukar S, Thomas KV, Mueller JF (2020c) Surveillance of SARS-CoV-2 RNA in wastewater: methods optimization and quality control are crucial for generating reliable public health information. Curr Opin Enviro Sci Health Environ Health: COVID-19 17:82–93. https://doi.org/10.1016/j.coesh.2020c.09.003
Ascoli JJ (2021) Could mutations of SARS-CoV-2 suppress diagnostic detection? Nat Biotechnol 39:270–274. https://doi.org/10.1038/s41587-021-00834-6
Balboa S, Mauricio-Iglesias M, Rodríguez S, Martínez-Lamas L, Vasallo FJ, Regueiro B, Lema JM (2020) The fate of SARS-CoV-2 in wastewater treatment plants points out the sludge line as a suitable spot for incidence monitoring (preprint). Epidemiology. https://doi.org/10.1101/2020.05.25.20112706
Bar-Or I, Weil M, Indenbaum V, Bucris E, Bar-Ilan D, Elul M, Levi N, Aguvaev I, Cohen Z, Shirazi R, Erster O, Sela-Brown A, Sofer D, Mor O, Mendelson E, Zuckerman NS (2021) Detection of SARS-CoV-2 variants by genomic analysis of wastewater samples in Israel. Sci Total Environ 789:148002. https://doi.org/10.1016/j.scitotenv.2021.148002
Berestycki H, Desjardins B, Heintz B, Oury J-M (2021) The effects of heterogeneity and stochastic variability of behaviours on the intrinsic dynamics of epidemics. medRxiv 2021.03.26.21254414. https://doi.org/10.1101/2021.03.26.21254414
Bressy A, Carré C, Caupos É, de Gouvello B, Deroubaix J-F, Deutsch J-C, Mailler R, Marconi A, Neveu P, Paulic L, Pichon S, Rocher V, Severin I, Soyer M, Moilleron R (2016) Cosmet’eau—Changes in the personal care product consumption practices: from whistle-blowers to impacts on aquatic environments. Environ Sci Pollut Res 23:13581–13584. https://doi.org/10.1007/s11356-016-6794-y
Buenestado-Serrano S, Recio R, Sola Campoy PJ, Catalán P, Folgueira MD, Villa J, Muñoz Gallego I, de la Cueva VM, Meléndez MA, Andrés Zayas C, Losa-García JE, Goyanes MJ, Fraile Torres A, Von Wermitz A, Fradejas-Villajos I, del Arco C, Campelo-Gutiérrez C, González Bodi S, López-Wolf D, Iglesias-Franco H, Pérez-Lago L, Arce Arnaez A, Rodriguez Baena E, Ordobas Gavin M, Muñoz P, Delgado R, Cardeñoso L, Viedma E, García de Viedma D (2021) First confirmation of importation and transmission in Spain of the newly identified SARS-CoV-2 B.1.1.7 variant. Enfermedades Infecciosas y Microbiología Clínica S0213–005X(21):00046–X. https://doi.org/10.1016/j.eimc.2021.02.006
Cao Y, Raith MR, Griffith JF (2015) Droplet digital PCR for simultaneous quantification of general and human-associated fecal indicators for water quality assessment. Water Res 70:337–349. https://doi.org/10.1016/j.watres.2014.12.008
Carcereny A, Garcia-Pedemonte D, Martínez-Velázquez A, Quer J, Garcia-Cehic D, Gregori J, Antón A, Andrés C, Pumarola T, Chacón-Villanueva C, Borrego CM, Bosch A, Guix S, Pintó RM (2022) Dynamics of SARS-CoV-2 Alpha (B.1.1.7) variant spread: the wastewater surveillance approach. Environ Res 208:112720. https://doi.org/10.1016/j.envres.2022.112720
Gasperi J, Garnaud S, Rocher V, Moilleron R (2008) Priority pollutants in wastewater and combined sewer overflow. Sci Total Environ 407:263–272. https://doi.org/10.1016/j.scitotenv.2008.08.015
Giacobbo A, Rodrigues MAS, Zoppas Ferreira J, Bernardes AM, de Pinho MN (2021) A critical review on SARS-CoV-2 infectivity in water and wastewater. What do we know? SciTotal Environ 774:145721. https://doi.org/10.1016/j.scitotenv.2021.145721
Hart OE, Halden RU (2020) Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: feasibility, economy, opportunities and challenges. Sci Total Environ 730:138875. https://doi.org/10.1016/j.scitotenv.2020.138875
Heijnen L, Elsinga G, de Graaf M, Molenkamp R, Koopmans MPG, Medema G (2021) Droplet digital RT-PCR to detect SARS-CoV-2 signature mutations of variants of concern in wastewater. Sci Total Environ 799:149456. https://doi.org/10.1016/j.scitotenv.2021.149456
Hong P-Y, Rachmdi AT, Mantilla-Calderon D, Alkahtani M, Bashawri YM, Al Qarni H, M.O’Reilly K, Zhou J (2021) Estimating the minimum number of SARS-CoV-2 infected cases needed to detect viral RNA in wastewater: to what extent of the outbreak can surveillance of wastewater tell us? Environ Res 195:110748. https://doi.org/10.1016/j.envres.2021.110748
Jafferali MH, Khatami K, Atasoy M, Birgersson M, Williams C, Cetecioglu Z (2021) Benchmarking virus concentration methods for quantification of SARS-CoV-2 in raw wastewater. Sci Total Environ 755:142939. https://doi.org/10.1016/j.scitotenv.2020.142939
Kitamura K, Sadamasu K, Muramatsu M, Yoshida H (2021) Efficient detection of SARS-CoV-2 RNA in the solid fraction of wastewater. Sci Total Environ 763:144587. https://doi.org/10.1016/j.scitotenv.2020.144587
Kocacemi BAK, Halil K, Ahmet S, Fahriye S, Ahmet Mete S, Bekir P (2020) SARS-CoV-2 detection in Istanbul wastewater treatment plant sludges. MedRxiv 11. https://doi.org/10.1101/2020.05.12.20099358v1
Kumar M, Kuroda K, Patel AK, Patel N, Bhattacharya P, Joshi M, Joshi CG (2021a) Decay of SARS-CoV-2 RNA along the wastewater treatment outfitted with Upflow Anaerobic Sludge Blanket (UASB) system evaluated through two sample concentration techniques. Sci Total Environ 754:142329. https://doi.org/10.1016/j.scitotenv.2020.142329
Kumar S, Singh R, Kumari N, Karmakar S, Behera M, Siddiqui AJ, Rajput VD, Minkina T, Bauddh K, Kumar N (2021b) Current understanding of the influence of environmental factors on SARS-CoV-2 transmission, persistence, and infectivity. Environ Sci Pollut Res 28:6267–6288. https://doi.org/10.1007/s11356-020-12165-1
Li B, Di DYW, Saingam P, Jeon MK, Yan T (2021) Fine-scale temporal dynamics of SARS-CoV-2 RNA abundance in wastewater during a COVID-19 lockdown. Water Res 197:117093. https://doi.org/10.1016/j.watres.2021.117093
Lu D, Huang Z, Luo J, Zhang X, Sha S (2020) Primary concentration – the critical step in implementing the wastewater based epidemiology for the COVID-19 pandemic: a mini-review. Sci Total Environ 747:141245. https://doi.org/10.1016/j.scitotenv.2020.141245
Mailler R, Mèche P, Rocher V (2021) What removals of pathogen indicators can be expected within large-scale wastewater treatment facilities in the context of wastewater reuse in Paris conurbation? Water Sci Technol 83(4):781–791. https://doi.org/10.2166/wst.2021.004
Medema G, Heijnen L, Brouwer A, Italiaander R, Elsinga G (2020) Presence of SARS-Coronavirus‑2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ Sci Technol Lett 7(7):511–516. https://doi.org/10.1021/acs.estlett.0c00357
Montier O, Lopez-Viveros M, Le Tallec X, Lacroix S, Tournié M, Soyeux E, Azimi S, Rocher V (2021) Comportement de l’ARN du SARS-CoV-2 au sein des filières de traitemetn des eaux et des boues du site Seine Valenton-SIAAP-SIVAL
Peterson SW, Lidder R, Daigle J, Wonitowy Q, Dueck C, Nagasawa A, Mulvey MR, Mangat CS (2022) RT-qPCR detection of SARS-CoV-2 mutations S 69–70 del, S N501Y and N D3L associated with variants of concern in Canadian wastewater samples. Sci Total Environ 810:151283. https://doi.org/10.1016/j.scitotenv.2021.151283
Rački N, Morisset D, Gutierrez-Aguirre I, Ravnikar M (2014) One-step RT-droplet digital PCR: a breakthrough in the quantification of waterborne RNA viruses. Anal Bioanal Chem 406:661–667. https://doi.org/10.1007/s00216-013-7476-y
Randazzo W, Truchado P, Cuevas-Ferrando E, Simón P, Allende A, Sánchez G (2020) SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res 181:115942. https://doi.org/10.1016/j.watres.2020.115942
Rocher V, Azimi S (2016) Qualité microbiologique des eaux en agglomération parisienne, Editions Johanet
Serra-Compte A, González S, Arnaldos M, Berlendis S, Courtois S, Loret JF, Schlosser O, Yáñez AM, Soria-Soria E, Fittipaldi M, Saucedo G, Pinar-Méndez A, Paraira M, Galofré B, Lema JM, Balboa S, Mauricio-Iglesias M, Bosch A, Pintó RM, Bertrand I, Gantzer C, Montero C, Litrico X (2021) Elimination of SARS-CoV-2 along wastewater and sludge treatment processes. Water Res 202:117435. https://doi.org/10.1016/j.watres.2021.117435
Singh M, Chazal M, Quarato P, Bourdon L, Malabat C, Vallet T, Vignuzzi M, van der Werf S, Behillil S, Donati F, Sauvonnet N, Nigro G, Bourgine M, Jouvenet N, Cecere G (2021) A virus-encoded microRNA contributes to evade innate immune response during SARS-CoV-2 infection (preprint). Mol Biol. https://doi.org/10.1101/2021.09.09.459577
Staley ZR, Boyd RJ, Shum P, Edge TA (2018) Microbial source tracking using quantitative and digital PCR to identify sources of fecal contamination in stormwater, river water, and beach water in a Great Lakes Area of Concern. Appl Environ Microbiol 84. https://doi.org/10.1128/AEM.01634-18
WHO (2020) Status of environmental surveillance for SARS-CoV-2 virus
Wurtzer S, Marechal V, Mouchel J-M, Maday Y, Teyssou R, Richard E, Almayrac JL, Moulin L (2020) Evaluation of lockdown impact on SARS-CoV-2 dynamics through viral genome quantification in Paris wastewaters (preprint). Epidemiology. https://doi.org/10.1101/2020.04.12.20062679
Wurtzer S, Waldman P, Levert M, Cluzel N, Almayrac JL, Charpentier C, Masnada S, Gillon-Ritz M, Mouchel JM, Maday Y, Boni M, Marechal V, Moulin L (2022) SARS-CoV-2 genome quantification in wastewaters at regional and city scale allows precise monitoring of the whole outbreaks dynamics and variants spreading in the population. Sci Total Environ 810:152213. https://doi.org/10.1016/j.scitotenv.2021.152213
Yaniv K, Ozer E, Shagan M, Lakkakula S, Plotkin N, Bhandarkar NS, Kushmaro A (2021) Direct RT-qPCR assay for SARS-CoV-2 variants of concern (Alpha, B.1.1.7 and Beta, B.1.351) detection and quantification in wastewater. Environ Res 201:111653. https://doi.org/10.1016/j.envres.2021a.111653
Yaniv K, Shagan M, Lewis YE, Kramarsky-Winter E, Weil M, Indenbaum V, Elul M, Erster O, Brown AS, Mendelson E, Mannasse B, Shirazi R, Lakkakula S, Miron O, Rinott E, Baibich RG, Bigler I, Malul M, Rishti R, Brenner A, Friedler E, Gilboa Y, Sabach S, Alfiya Y, Cheruti U, Nadav D, Moran-Gilad J, Berchenko Y, Bar-Or I, Kushmaro A (2021) City-level SARS-CoV-2 sewage surveillance. Chemosphere 283:131194. https://doi.org/10.1016/j.chemosphere.2021.131194
Acknowledgements
The authors would like to acknowledge Veronique Bremont and Jennifer Mas for their implication on sampling as well the SIAAP teams that participated to sampling and analyses, including SEV, SEG, SEM, and the SEC SIAAP central laboratory.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Experimental and data collection were performed by Franz Durandet and Elodie Pichon, while data analysis was done by Melissa Lopez Viveros, Sam Azimi, and Vincent Rocher. The first draft of the manuscript was written by Melissa Lopez Viveros, and all authors commented on previous versions of the manuscript. Sam Azimi and Vincent Rocher were in charge of supervision and validation. Celine Roose-Amsaleg and Ariane Bize greatly contributed to final review and editing. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
Not applicable.
Additional information
Responsible Editor: Philippe Garrigues
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Viveros, M.L., Azimi, S., Pichon, E. et al. Wild type and variants of SARS-COV-2 in Parisian sewage: presence in raw water and through processes in wastewater treatment plants. Environ Sci Pollut Res 29, 67442–67449 (2022). https://doi.org/10.1007/s11356-022-22665-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-022-22665-x