Abstract
RNA silencing is a form of nucleic-acid-based immunity, targeting viruses and genomic repeated sequences. First documented in plants and invertebrate animals, this host defence has recently been identified in mammals. RNAi is viewed as a conserved ancient mechanism protecting genomes from nucleic acid invaders. However, these tamed sequences are known to occasionally escape this host surveillance and invade the genome of their host. This response is consistent with the overall idea that parasitic sequences compete with cells to systematically counter host defences. Using examples taken from the current literature, we illustrate the dynamic move–countermove game played between these two protagonists, the host cell and its parasitic sequences, and discuss the consequences of this game on genome stability.
Similar content being viewed by others
Introduction
Eukaryotic genomes are largely composed of repetitive DNA sequences. Among them, transposable elements have the potential to perform replication cycles involving DNA or RNA intermediates. These repeated and mobile sequences have been found in all living organisms, and can comprise up to 40% of the genome, as is the case in humans. Tight control of these invaders is thus an important feature of their regulation to prevent eukaryotic genomes from their mutational threat. During the last few years, evidence has emerged that the host has developed mechanisms to silence such repeated sequences or destroy any foreign genetic material detected in a cell. Several mechanisms have been implicated, but one of them recently emerged as one of the main actors of this regulation. It is a post-transcriptional mechanism called the RNA interference pathway, or RNAi, which acts as a sequence-specific RNA degradation mechanism.
The RNAi pathway
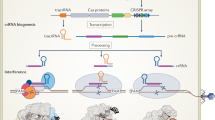
Gene silencing by a post-transcriptional mechanism involving homologous double-stranded RNA was first discovered in artificial systems where double-strand RNA (dsRNA) was introduced by injection or by expression of transgenic constructs. Since then, many genetic and biochemical studies have furthered our understanding of the mechanism by which RNAi acts. RNAi is usually seen as a two-step reaction (Figure 1). In the first step, a long dsRNA is processed into short (21–25 nucleotides) small interfering RNAs (siRNAs) (Hannon, 2002). siRNAs are subsequently incorporated into a silencing complex, called RISC (RNAi-Induced Silencing Complex), where they serve as templates to guide the endonucleolytic cleavage of homologous mRNAs (Hammond et al, 2001). The first step is performed by a family of ribonucleases, the RNAse III family, and more particularly an enzyme called Dicer. Dicer is able to recognize and cleave long dsRNA molecules to generate the siRNAs. Unlike many animals, Drosophila or plant genomes encode several Dicer or Dicer-like proteins, indicating that these enzymes could play a more complex role than that already described (Xie et al, 2004). For example, Dcr-2, one of the two Dicer proteins encoded by Drosophila melanogaster, cleaves dsRNAs into 21 nucleotide dsRNAs comprising a core of 19 nucleotides long dsRNAs and one sorting nucleotide on each side. However, Dcr-2 is also required at a latter step of the RNAi pathway in an initiator complex that binds siRNAs and loads them onto the RISC (Liu et al, 2003).
Mechanisms of RNA silencing in Drosophila melanogaster. (a) RNAi pathway and mRNA destruction. The RNAi pathway is initiated by the recognition of double-stranded RNAs by Dicer 2. This enzyme cleaves them to form short double-strand RNAs called siRNAs. The heterodimer Dicer2–R2D2 targets siRNAs to a first ribonucleoproteic complex called RISC (RNAi-induced silencing complex). This RISC matures and gives rise to an effector complex called holo-RISC with only one strand of the siRNA. This siRNA is then used as a template to hybridize and target a homologous mRNA for destruction. (b) miRNAs and RNA silencing. miRNA precursors, called pre-miRNAs, are encoded by host genomes. They are cleaved a first time by an enzyme called Drosha that acts in the nucleus as a multiprotein complex called the Microprocessor complex. The processed pre-miRNAs are then exported to the cytoplasm. Dicer1 recognizes the pre-miRNAs and cleaves them a second time to give 21-bp RNAs that will either direct the translational repression of homologous mRNAs or target mRNAs for destruction.
The second step of the RNAi pathway is the formation of the RISC complex, a large ribonucleoprotein complex considered to be a homology-dependent endonuclease, which seeks out and destroys the messenger RNA. Members of the argonaute protein family are core components of these RISC or RISC-like complexes. They are genetically required for RNA silencing in each organism where their function has been studied, but the exact role of this family has generally not been determined. Lots of proteins acting in the RNAi pathway share a common domain of activity, that is the DEA(H/D)-box helicase, but no specific biochemical function in RNAi has been ascribed to these helicases. The main catalytic activity of the RISC complex is to cut target RNA. In humans and mice, this is performed by an enzyme called Ago2 that belongs to the argonaute protein family (Liu et al, 2004). In Drosophila, two nucleases have been isolated in the RISC: DICER, and Tudor SN. However, which nuclease slices the target RNA remains unknown (Caudy et al, 2003; Schwarz et al, 2004).
Pham et al (2004) have shown that three distinct RISC complexes associated with siRNAs in cell extracts exist in Drosophila. A first complex associates directly with the initiator complex containing Dcr-2, and serves as a precursor to the other two complexes. The second complex leads to the formation of the third complex that completes all the maturation steps to become a holo-RISC. One of the roles of the RISC is to unwind the double-stranded siRNA into a single-stranded template RNA. Thermodynamic and biochemical studies have shown that small changes in siRNA sequence have profound effects on which strand of the siRNA is incorporated into the RISC. This led to the conclusion that RISC assembly is governed by an enzyme that selects the strand incorporated in the RISC that may lead to a structural and functional asymmetry of certain siRNAs. It has been proposed that siRNA structures assign one strand to enter the RISC and the other strand to be destroyed (Khvorova et al, 2003; Schwarz et al, 2003). Tomari et al (2004) have shown that in Drosophila, the orientation of the Dicer2/R2D2 protein heterodimer on the siRNA duplex determines which siRNA strand associates with the core RISC protein.
miRNAs are involved in the post-transcriptional silencing pathway
Another family of small RNA molecules able to silence genes was discovered in the 1990s. These RNAs, called microRNAs (miRNAs), are small RNA molecules 21 nucleotides long, with a very similar structure to siRNAs, but are single-stranded. They come from the cleavage by Dicer of stem loop precursor RNAs (pre-miRNAs) encoded within the genomes of plants and animals (He and Hannon, 2004). The miRNAs act as components of ribonucleoproteic complexes regulating gene expression through interactions with their target RNAs. Target genes are often genes involved in regulating key developmental events. Despite this common structure, plant and animal miRNAs exert their control in fundamentally different ways (Millar and Waterhouse, 2005). Generally speaking, animal miRNAs repress gene expression by mediating translational silencing through multiple interactions in the 3′UTR of the target mRNA. However, miRNAs can also inhibit mRNA expression by similar mechanisms to siRNAs. For instance, plant miRNAs often regulate their target genes by directing mRNA cleavage at single sites in the coding regions. The choice between translational repression or cleavage of target RNA is governed by the homology between the small noncoding RNA and the target. An imperfect complementarity leads to translational repression, whereas a fully complementary site will be hybridized and cleaved (Doench et al, 2003; Zeng et al, 2003).
Drosha, a nuclear RNAse III acting as part of a multiprotein complex called the microprocessor complex, initiates the miRNA processing by cutting the stem-loop precursor RNA within the nucleus (Lee et al, 2003; Denli et al, 2004; Gregory et al, 2004). The processed pre-miRNA is then specifically recognized by a nuclear export factor termed Exportin 5, which transports the pre-miRNA to the cytoplasm (Lund et al, 2004). There, the pre-miRNA is cleaved by Dicer to give a ∼21-bp RNA duplex intermediate, one strand of which is selectively incorporated into the RISC. In Drosophila, Dcr-1 matures miRNAs, and Dcr-2 matures siRNAs. Does this mean that these small RNAs are incorporated in different effector complexes? The fact that only a fraction of these complexes is associated with ribosomes indicates that specificities may exist among the small RNAs, but these putative specificities remain to be characterized.
Whereas miRNAs are expressed in a developmental stage or tissue-specific manner and are believed to play a key role in gene regulation; their role in the defence of host genomes against viruses and transposons has only recently been established. It has been shown that a miRNA synthesized by the mouse genome targets the RNA genome of PFV (Primate Foamy Virus) and leads to its destruction (Lecellier et al, 2005). Given the presumed importance of such an innate antiviral defence mechanism driven by miRNAs, it is reasonable to hypothesize that other miRNA encoded by the host and targeting exogenous invaders will be discovered in a wide range of organisms, if not all. However, it should be added that some viral genomes also have the potential to encode miRNAs that are able to interfere with their host genome, which suggests a complex role of the RNA-silencing mechanisms in host/pathogen relationships (Pfeffer et al, 2004; Sullivan et al, 2005).
Strategies for efficiency
If used as a first-line defence against viruses and transposable elements, RNAi has to destroy large amounts of targeted RNA very efficiently, whatever the affected cells. Strategies to increase RNAi-silencing efficiency have been elucidated in two types of organisms, plants and Caenorhabditis elegans. Two strategies, transitivity and systemy, have been characterized (Figure 2).
Mechanisms for transitivity and systemy. In cell (a), double-stranded RNAs are recognized by the RNAi pathway and cleaved by a Dicer-like protein to form siRNAs. These siRNAs seek out and destroy homologous target RNAs in the cell where they have been generated. This RNA silencing can spread over 2–5 cells (indicated as (b) cells) by diffusing with the help of a transmembrane protein called SID. This phenomenon is called systemy. As illustrated (but still controversial), the spreading molecules may be siRNAs themselves. A mobile signal allows the RNAi-silencing pathway to be activated in neighbouring cells (indicated as (c)) and to target the same mRNA. In some organisms, an enzyme called RdRp generates new siRNAs able to reinitiate RNAi and amplify the system (see cell (c)). This phenomenon is called transitivity.
Transitivity: amplification of the signal
The first strategy employed to improve RNAi efficiency is to amplify the amount of the key molecules acting in RNAi, ie siRNAs. This amplification mechanism, called transitivity, means that siRNA bearing new homology sequences upstream or downstream of the target zone are synthesized. These new siRNAs can then seek out and destroy any homologous mRNA. Initial studies performed in plants and nematodes have reported that RNA-dependent RNA polymerases or RdRp could be involved in the generation of secondary siRNAs homologous to the full-length of the target RNA (Smardon et al, 2000; Forrest et al, 2004). Two models of action have been proposed. First, RdRp is able to generate newly synthesized siRNAs. Second, RdRp uses siRNA as a primer for extension, and synthesizes new long dsRNAs (Makeyev and Bamford, 2002). These newly synthesized RNAs are able to reinitiate RNAi and thus amplify the system. A difference in the length of siRNAs has been observed: primary siRNAs are 21–22 nucleotides long, whereas siRNAs resulting from an amplification step are 24–26 long. These discoveries highlighted an unexpected complexity of small RNA molecules involved in the RNAi pathway (Hamilton et al, 2002).
This begs the question of why transitivity has only been characterized in plants and nematodes to date. Numerous observations suggest that an RdRp activity is not be involved in RNAi pathway of some organisms. Several data support this assumption in Drosophila. First, no member of the RdRp family has been found in the Drosophila genome. Second, the integrity of the 3′ hydroxyl group, necessary for the RdRp to be active, is not required for the RNAi pathway studied in Drosophila embryo lysates (Schwarz et al, 2002). Third, when siRNAs were used to knock down a gene, Zamore et al (2000) did not find any evidence of cleavage in the RNA sequence located upstream of the siRNA-targeted region. Finally, Roignant et al (2003) showed that no trans-silencing of a GFP transgene was observed when initiating RNAi against a fusion GFP-target transgene. Nevertheless, there is evidence to suggest that transitivity may indeed exist in Drosophila. For instance, Lipardi et al (2001) provided evidence for an RdRp activity in Drosophila embryonic extracts. Additionally, siRNAs homologous to the entire silenced gene can be generated in Drosophila in the course of cosuppression, a mechanism able to silence genes present in multicopies and initiate an RNAi-related silencing (Pal-Bhadra et al, 2002). Overall, the possibility that transitivity exists in Drosophila or other organisms has not been excluded, but transitivity may have been rarely detected due to its high specificity for certain target genes, or for any steps in the pathway.
Systemy: the mobile silencing signal
Another strategy used by species to improve their silencing efficiency is based on the fact that RNAi is not cell-autonomous. In both C. elegans and plants, locally initiated silencing can spread to distant sites throughout the organism. This indicates that an as yet unidentified mobile silencing signal exists that cooperates with RNAi. In nematodes, this mobile signal can travel long distances via cell-to-cell movements. Several proteins required for this trafficking have been described, including the multispan transmembrane protein SID-1 (Duxbury et al, 2005), or Rsd-3, a homolog of the human enthoprotin, which was previously shown to be involved in vesicle-trafficking (Tijsterman et al, 2004). In plants, two types of RNA-silencing spread exist: a cell-to-cell movement, very similar to the nematode one, and a long-range spreading. Indirect evidence indicates that RNA-silencing moves over long distances through the phloem and activates silencing in cells by spreading through plasmodesmata. Himber et al (2003) reported that the mobile signal is able to spread to nearby cells using specific proteins, where it can activate RNAi and, if an amplification system is present, reinitiate a local cell-to-cell movement. While genes required for an active systemy have been partly identified, the mobile signal involved remains to be determined. siRNAs are very good candidates, as they are always associated with RNA silencing and have the ability to reinitiate RNAi. However, mutants in plants and worms defective for siRNA accumulation or production are not defective for systemic silencing. Other candidates could be the long dsRNAs themselves, or the targets bearing defects or tagged by proteins. In all these models, the fact that the same species exhibiting systemy also show transitivity suggests that a link exists between these mechanisms.
Counteracting the RNAi control
The hypothesis that RNAi can form an important part of the innate response was first supported by the observation that many plant viruses encode proteins that inhibit RNAi (Voinnet et al, 1999). For example, tomato bushy tombusvirus and turnip mosaic virus have been reported to encode viral suppressors of RNA silencing which reduce cellular antiviral effects so that the respective viruses can accumulate to high titers (Vance and Vaucheret, 2001; Zamore, 2004; Dunoyer and Voinnet, 2005). Although the earliest studies were performed on plant viruses, suppressors that can inhibit some aspects of the RNAi response have also been identified in animal viruses. The B2 protein encoded by the ubiquitous flock house virus (FHV), which is an RNA virus that can replicate in both plant and insect cells, has been shown to block the ability of cells to mount an FHV-specific RNAi response (Li et al, 2002). The influenza virus can inhibit RNAi when expressed in Drosophila S2 cells (Li et al, 2004). Very recently, it has been proved that vertebrate viruses have evolved suppressors of RNA silencing to escape a nucleic-acid-based cellular defence, present in their host. The protein Tas encoded by the PFV (primate foamy virus) suppresses microRNA-directed functions in mammalian cells and displays cross-kingdom antisilencing activities (Lecellier et al, 2005). The Tat protein encoded by HIV-1 has been recently shown to neutralize the cell's RNA-silencing defence (Bennasser et al, 2005). Furthermore, Adenovirus VA1 noncoding RNA can inhibit siRNA and miRNA biogenesis in human cells (Lu and Cullen, 2004).
The diversity of the suppressors described so far is correlated with diversity in their mechanisms of action. For instance, HC-PRO, a silencing suppressor encoded by potyviruses, is able to reverse silencing by affecting siRNA accumulation without reversing the mobile signal (Mette et al, 2001); Cmv2b, a protein encoded by PVX (Potato Virus X), interferes with mobile silencing signal transport (Brigneti et al, 1998); P19 encoded by the tombusvirus cancels out the silencing pathway by binding and sequestering siRNAs (Vargason et al, 2003). In addition to binding siRNAs, P19 also binds miRNAs, and has crosseffects with the miRNA pathway. As a final example, Tat encoded by HIV-1 subverts the ability of Dicer to process precursor double-stranded RNAs into siRNAs (Bennasser et al, 2005).
As the whole-gene regulation of the cell is disrupted by virus-encoded silencing suppressors, it is hard to estimate the side effects of such infections. For example, we recently found that PFV-Tas may inhibit the post-transcriptional silencing exerted on a Drosophila endogenous retrotransposon called ZAM (unpublished results). Our results indicate that upon an infection, viral suppressors may release the silencing imprinted on endogenous repeated elements, and therefore contribute to disrupting genome integrity. It follows that the targets of viral products in their hosts should nowadays be investigated in detail, to understand their potential deleterious impact on the cell biology.
Finally, it is important to point out that genomes themselves encode endogenous silencing suppressors (Kennedy et al, 2004). Such endogenous negative regulators of RNAi may have a crucial role in controlling the RNAi pathway, and may explain the reversible, impermanent nature of this defence.
Connection between RNAi and transcriptional silencing
As part of the silencing machinery acting in the cell against nucleic invaders, RNAi plays a major role and acts as one of the tools employed by the cell to protect itself against their mutagenic impact. However, gene silencing as a defence against transposons and viruses also relies on another mechanism acting at the transcriptional level – transcriptional gene silencing or TGS. In plants, TGS occurs when promoter homology exists among transgenes or with endogenous genes. Until recently, both RNAi and TGS were considered as separate pathways, with TGS acting at the transcriptional level and RNAi acting on RNA metabolism. Recently, a convergence of observations from diverse experimental systems suggested that a conserved mechanism might link both homology-dependent gene-silencing responses.
TGS is often thought to involve a local chromatin modification whereby DNA and histones are chemically modified to recruit proteins inhibiting transcriptional activity and condensing chromatin. Hallmarks of TGS are DNA cytosine methylation (except in flies) and histone three lysine 9 methylation (H3K9met). The discovery that mutations in several genes affecting RNAi in C. elegans affect the regulation of transposons suggested that TGS and RNAi silencing could form alternative, nonexclusive pathways to regulate the same elements. The major breakthrough in the distinction between TGS and post-transcriptional gene silencing (PTGS) or RNAi came from the discovery that viruses and transgenes encoding dsRNA silence homologous transgenes by inducing either TGS or RNAi, depending on whether they share homology with its promoter or its protein encoding sequence, respectively. In plants, the introduction of dsRNA initiates a de novo methylation of homologous DNA called RNA-dependent DNA-methylation (RdDM) (Vaucheret and Fagard, 2001). This methylation only alters gene expression when occurring near the promoter. In addition to enlightening a direct link between TGS and PTGS, this result indicates that RNA could be the main actor targeting DNA sequences for TGS. Further data have come from the study of a gene called Superman in Arabidopsis. This gene undergoes an epigenetic shut-off acting at the transcriptional level, called the clark kent allele (Jacobsen and Meyerowitz, 1997). Ago1 mutants were shown to affect the establishment of the silent state of clark kent, thus directly implicating RNAi in TGS. The observation that RNAi-related mechanisms affect both transcriptional and PTGS have also been made in Drosophila when studying the silencing affecting transgenes with the alcohol dehydrogenase (Adh) transcription unit (Pal-Bhadra et al, 2002). Pal-Bhadra et al (2002) showed that TGS is Polycomb-dependent and occurs when Adh is driven by the white eye color gene promoter. Conversely, full-length Adh transgenes are silenced post-transcriptionally with molecular hallmarks typical of RNAi. Moreover, mutations in piwi required for RNAi block PTGS and one aspect of TGS.
The underlying mechanism linking the TGS and RNAi pathways remains unclear. However, genetic studies performed in the yeast Schizosaccharomyces pombe have provided further insight (Figure 3). The repetitive DNA at centromeres is kept silent by H3K9 methylation and the binding of Swi6 to the modified chromatin. Volpe et al (2002) revealed that deletion of genes encoding the RNAi pathway leads to loss of gene silencing and entire heterochromatin disorganization. Overlapping transcripts initiated from outer repeat sequences present in this part of the genome are thought to allow dsRNA to form. They are processed into homologous siRNAs able to direct a new round of heterochromatinization. The existence of siRNAs homologous to centromeric repeats strengthen their model.
The expression of double-stranded RNA triggers the assembly of silent heterochromatin in Schizosaccharomyces pombe. Top and bottom strands of an open heterochromatin region are transcribed. These overlapping transcripts hybridize and form double-stranded RNA molecules homologous to the DNA segment. dsRNAs enter the RNAi pathway and are cleaved to siRNAs. The small RNAs are then incorporated into a new complex called RITS (RNAi-induced transcriptional silencing complex), which is able to target homologous DNA. The RITS recruits Clr4 that methylates lysine 9 of histone H3 at the target locus. This allows binding of Swi6, the mammalian HP1 homolog, on the modified chromatin, and enables the formation of silent heterochromatin able to spread along the chromatin fiber.
This RNAi-mediated heterochromatin assembly in fission yeast requires an RNAi effector complex, called the RNA-induced transcriptional silencing (RITS) complex, which contains Ago1 (argonaute homolog), Chp1 (a heterochromatin-associated chromodomain protein) and Tas3 (Verdel et al, 2004). Generation of these heterochromatic siRNA requires the action of RdRP that acts in a complex called RDRC. The combined action of RDRC and RITS together with their physical interaction enables the fission yeast to target heterochromatinization to repeat DNA (Shankaranarayana et al, 2003; Motamedi et al, 2004). In accordance with the idea that a link exists between TGS and RNAi, it was also shown in D. melanogaster that RNAi-defective mutants display a heterochromatin deregulation, and a loss of HP1 localization (Pal-Bhadra et al, 2004).
The finding that HP1 and Su(Var)3-9 may repress euchromatic genes supports the idea that chromatin modifications similar to those that operate over large areas in heterochromatin (eg pericentromeric heterochromatin) can act in a highly localized and targeted manner to silence transcription of euchromatic sequences. Aravin et al (2001) proposed that siRNA generated from heterochromatic regions mainly composed of transposable elements could target homologous sequences present in euchromatin regions, silence these elements, and cause the assembly of localized patches of silent chromatin (Aravin et al, 2001).
Host genome and nucleic invaders
In many organisms, transposable elements or their relics are packaged in silent chromatin. Data obtained by various groups have now linked naturally occurring transposon silencing to both heterochromatin and RNAi (Desset et al, 2003; Schramke and Allshire, 2003; Sarot et al, 2004). When an early search for natural targets of RNAi was performed, Baulcombe (2002) showed in plant models that RNAi works as a real antiviral defence system targeting viruses and endogenous repeated sequences. This was confirmed when a subset of mutations selected as defective for RNAi were shown to mobilize families of transposable elements. In D. melanogaster, mutations in spindle E, a gene involved in RNAi, led to a derepression of retrotransposons in the germline (Aravin et al, 2004), and the argonaute protein piwi is required for this repression in the male germline (Kalmykova et al, 2005). Moreover, it has recently been shown that gypsy and ZAM, two endogenous retroviruses of D. melanogaster, are regulated by a piwi-dependent mechanism in somatic tissues (Sarot et al, 2004) (and our unpublished results). In Trypanosoma brucei, Ago1 deficiency leads to an increase in retroposon transcript abundance. In fungi, such as Neurospora crassa, transposons are known to be repressed by repeat-induced point mutation (RIP). Chicas et al (2004) showed in these organisms that RNAi is able to target transposon RNAs and act in concert with RIP to silence them.
Plasterk and colleagues elucidated a mechanism whereby fortuitous and aberrant transcription of dispersed Tc1 copies can lead to dsRNA formation as a result of a ‘snap back’ of the terminal inverted repeats which sets RNAi against this transposon (Sijen and Plasterk, 2003). Recently, cloning and characterization of a naturally occurring locus able to heritably silence the otherwise highly active MuDR transposon in maize was reported (Slotkin et al, 2005). This locus, called Mu killer (Muk), results from the inverted duplication of a partially deleted autonomous MuDR element located at the breakpoint of a genomic deletion. Muk produces a hybrid hairpin transcript that is processed into small RNAs, which are then amplified when target MuDR transcript is present. This study provides the first example of a naturally occurring transposon derivative capable of initiating the heritable silencing of an active transposon family.
The existence of such an RNA-based defence mechanism against invading elements could be particularly important for organisms such as plants and invertebrate animals which lack protein-based adaptive immunity. Nevertheless, evidence has recently emerged that mammals have also developed post-transcriptional silencing to silence or destroy foreign genetic material detected in a cell. In preimplantation mouse embryos, inhibition of the RNAi pathway resulted in a 50% increase in both murine endogenous retrovirus-L (MuERV-L) and intracisternal A particle (IAP) transcript abundance (Svoboda et al, 2004). In human cells, Lecellier et al (2005) have shown that RNA-silencing limits the replication of at least one mammalian virus, PFV. Furthermore, Bennasser et al (2005) demonstrated that natural HIV-1 infection also provokes nucleic-acid-based immunity. Therefore, if this ancestral genomic defence is a general mechanism targeting nucleic invaders from plant to mammalian genomes, it can be suspected that numerous miRNA encoded by all the organisms remain to be discovered. It can therefore be hypothesized that amongst the hundreds of conserved and nonconserved human miRNAs recently reported by Bentwich et al (2005), some of them will revealed to be essential actors of viral and repeated sequence silencing.
Conclusion
Overall, data obtained these last few years indicate that RNAi acts as a sequence-specific silencing mechanism which provides an ancestral cellular defence present in a wide range of organisms from plants to humans, and which is required to inactivate exogenous viruses or genes present in multiple copies within the genome. It is now clear that a connection exists between transcriptional gene silencing and post-transcriptional silencing to constrain the expression of these invaders. Further data will help to understand how, in concert, they contribute to preserving genome integrity at the various stages of development.
Another challenge in this field will be to fully dissect the mechanisms employed by numerous viruses or transposable elements to circumvent this cellular defence, and the exact role of silencing suppressors. A greater understanding of this complex relationship between parasitic elements and their host will compellingly illustrate the dynamic evolutionary ‘cat and mouse’ game played within the cells.
References
Aravin AA, Klenov MS, Vagin VV, Bantignies F, Cavalli G, Gvozdev VA (2004). Dissection of a natural RNA silencing process in the Drosophila melanogaster germ line. Mol Cell Biol 24: 6742–6750.
Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA (2001). Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol 11: 1017–1027.
Baulcombe D (2002). Viral suppression of systemic silencing. Trends Microbiol 10: 306–308.
Bennasser Y, Le SY, Benkirane M, Jeang KT (2005). Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity 22: 607–619.
Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O et al (2005). Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet 37: 766–770.
Brigneti G, Voinnet O, Li WX, Ji LH, Ding SW, Baulcombe DC (1998). Viral pathogenicity determinants are suppressors of transgene silencing in nicotiana benthamiana. Embo J 17: 6739–6746.
Caudy AA, Ketting RF, Hammond SM, Denli AM, Bathoorn AM, Tops BB et al (2003). A micrococcal nuclease homologue in RNAi effector complexes. Nature 425: 411–414.
Chicas A, Cogoni C, Macino G (2004). RNAi-dependent and RNAi-independent mechanisms contribute to the silencing of RIPed sequences in Neurospora crassa. Nucleic Acids Res 32: 4237–4243.
Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ (2004). Processing of primary microRNAs by the microprocessor complex. Nature 432: 231–235.
Desset S, Meignin C, Dastugue B, Vaury C (2003). COM, a heterochromatic locus governing the control of independent endogenous retroviruses from Drosophila melanogaster. Genetics 164: 501–509.
Doench JG, Petersen CP, Sharp PA (2003). siRNAs can function as miRNAs. Genes Dev 17: 438–442.
Dunoyer P, Voinnet O (2005). The complex interplay between plant viruses and host RNA-silencing pathways. Curr Opin Plant Biol 8: 415–423.
Duxbury MS, Ashley SW, Whang EE (2005). RNA interference: a mammalian SID-1 homologue enhances siRNA uptake and gene silencing efficacy in human cells. Biochem Biophys Res Commun 331: 459–463.
Forrest EC, Cogoni C, Macino G (2004). The RNA-dependent RNA polymerase, QDE-1, is a rate-limiting factor in post-transcriptional gene silencing in neurospora crassa. Nucleic Acids Res 32: 2123–2128.
Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N et al (2004). The Microprocessor complex mediates the genesis of microRNAs. Nature 432: 235–240.
Hamilton A, Voinnet O, Chappell L, Baulcombe D (2002). Two classes of short interfering RNA in RNA silencing. Embo J 21: 4671–4679.
Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ (2001). Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293: 1146–1150.
Hannon GJ (2002). RNA interference. Nature 418: 244–251.
He L, Hannon GJ (2004). MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5: 522–531.
Himber C, Dunoyer P, Moissiard G, Ritzenthaler C, Voinnet O (2003). Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. Embo J 22: 4523–4533.
Jacobsen SE, Meyerowitz EM (1997). Hypermethylated SUPERMAN epigenetic alleles in arabidopsis. Science 277: 1100–1103.
Kalmykova AI, Nurminsky DI, Ryzhov DV, Shevelyov YY (2005). Regulated chromatin domain comprising cluster of co-expressed genes in Drosophila melanogaster. Nucleic Acids Res 33: 1435–1444.
Kennedy S, Wang D, Ruvkun G (2004). A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature 427: 645–649.
Khvorova A, Reynolds A, Jayasena SD (2003). Functional siRNAs and miRNAs exhibit strand bias. Cell 115: 209–216.
Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C et al (2005). A cellular microRNA mediates antiviral defense in human cells. Science 308: 557–560.
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J et al (2003). The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415–419.
Li H, Li WX, Ding SW (2002). Induction and suppression of RNA silencing by an animal virus. Science 296: 1319–1321.
Li WX, Li H, Lu R, Li F, Dus M, Atkinson P et al (2004). Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc Natl Acad Sci USA 101: 1350–1355.
Lipardi C, Wei Q, Paterson BM (2001). RNAi as random degradative PCR: siRNA primers convert mRNA into dsRNAs that are degraded to generate new siRNAs. Cell 107: 297–307.
Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ et al (2004). Argonaute2 is the catalytic engine of mammalian RNAi. Science 305: 1437–1441.
Liu Q, Rand TA, Kalidas S, Du F, Kim HE, Smith DP et al (2003). R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 301: 1921–1925.
Lu S, Cullen BR (2004). Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis. J Virol 78: 12868–12876.
Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U (2004). Nuclear export of microRNA precursors. Science 303: 95–98.
Makeyev EV, Bamford DH (2002). Cellular RNA-dependent RNA polymerase involved in posttranscriptional gene silencing has two distinct activity modes. Mol Cell 10: 1417–1427.
Mette MF, Matzke AJ, Matzke MA (2001). Resistance of RNA-mediated TGS to HC-Pro, a viral suppressor of PTGS, suggests alternative pathways for dsRNA processing. Curr Biol 11: 1119–1123.
Millar AA, Waterhouse PM (2005). Plant and animal microRNAs: similarities and differences. Funct Integr Genomics 5: 129–135.
Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D (2004). Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 119: 789–802.
Pal-Bhadra M, Bhadra U, Birchler JA (2002). RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol Cell 9: 315–327.
Pal-Bhadra M, Leibovitch BA, Gandhi SG, Rao M, Bhadra U, Birchler JA et al (2004). Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303: 669–672.
Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J et al (2004). Identification of virus-encoded microRNAs. Science 304: 734–736.
Pham JW, Pellino JL, Lee YS, Carthew RW, Sontheimer EJ (2004). A Dicer-2-dependent 80s complex cleaves targeted mRNAs during RNAi in Drosophila. Cell 117: 83–94.
Roignant JY, Carre C, Mugat B, Szymczak D, Lepesant JA, Antoniewski C (2003). Absence of transitive and systemic pathways allows cell-specific and isoform-specific RNAi in Drosophila. Rna 9: 299–308.
Sarot E, Payen-Groschene G, Bucheton A, Pelisson A (2004). Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics 166: 1313–1321.
Schramke V, Allshire R (2003). Hairpin RNAs and retrotransposon LTRs effect RNAi and chromatin-based gene silencing. Science 301: 1069–1074.
Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD (2003). Asymmetry in the assembly of the RNAi enzyme complex. Cell 115: 199–208.
Schwarz DS, Hutvagner G, Haley B, Zamore PD (2002). Evidence that siRNAs function as guides, not primers, in the Drosophila and human RNAi pathways. Mol Cell 10: 537–548.
Schwarz DS, Tomari Y, Zamore PD (2004). The RNA-induced silencing complex is a Mg2+-dependent endonuclease. Curr Biol 14: 787–791.
Shankaranarayana GD, Motamedi MR, Moazed D, Grewal SI (2003). Sir2 regulates histone H3 lysine 9 methylation and heterochromatin assembly in fission yeast. Curr Biol 13: 1240–1246.
Sijen T, Plasterk RH (2003). Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature 426: 310–314.
Slotkin RK, Freeling M, Lisch D (2005). Heritable transposon silencing initiated by a naturally occurring transposon inverted duplication. Nat Genet 37: 641–644.
Smardon A, Spoerke JM, Stacey SC, Klein ME, Mackin N, Maine EM (2000). EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr Biol 10: 169–178.
Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D (2005). SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature 435: 682–686.
Svoboda P, Stein P, Anger M, Bernstein E, Hannon GJ, Schultz RM (2004). RNAi and expression of retrotransposons MuERV-L and IAP in preimplantation mouse embryos. Dev Biol 269: 276–285.
Tijsterman M, May RC, Simmer F, Okihara KL, Plasterk RH (2004). Genes required for systemic RNA interference in Caenorhabditis elegans. Curr Biol 14: 111–116.
Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD (2004). A protein sensor for siRNA asymmetry. Science 306: 1377–1380.
Vance V, Vaucheret H (2001). RNA silencing in plants – defense and counterdefense. Science 292: 2277–2280.
Vargason JM, Szittya G, Burgyan J, Tanaka Hall TM (2003). Size selective recognition of siRNA by an RNA silencing suppressor. Cell 115: 799–811.
Vaucheret H, Fagard M (2001). Transcriptional gene silencing in plants: targets, inducers and regulators. Trends Genet 17: 29–35.
Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI et al (2004). RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303: 672–676.
Voinnet O, Pinto YM, Baulcombe DC (1999). Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc Natl Acad Sci USA 96: 14147–14152.
Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA (2002). Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833–1837.
Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D et al (2004). Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2: E104.
Zamore PD (2004). Plant RNAi: how a viral silencing suppressor inactivates siRNA. Curr Biol 14: R198–200.
Zamore PD, Tuschl T, Sharp PA, Bartel DP (2000). RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101: 25–33.
Zeng Y, Yi R, Cullen BR (2003). MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc Natl Acad Sci U S A 100: 9779–9784.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Buchon, N., Vaury, C. RNAi: a defensive RNA-silencing against viruses and transposable elements. Heredity 96, 195–202 (2006). https://doi.org/10.1038/sj.hdy.6800789
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6800789