Abstract
The composition of the gut microbiome of patients with advanced non-small cell lung cancer is currently considered a factor influencing the effectiveness of treatment with immune checkpoint inhibitors. We aimed to evaluate the baseline gut microbiome composition in patients before receiving first-line immunotherapy alone or combined with chemoimmunotherapy. We performed 16S rRNA sequencing based on hypervariable regions. Stool samples were collected from 52 patients with advanced NSCLC treated with immunotherapy or chemoimmunotherapy before treatment. We found that the Ruminococcaceae family, species Alistipes sp. genus Eubacterium ventriosum group and genus Marvinbryantia may be intestinal, microbiological predictors of response to treatment. Genus Akkermansia and species from the [Clostridum] leptum group predicted the length of PFS (progression-free survival). Longer OS (overall survival) is associated with bacteria from the Ruminococcaceae family genera [Eubacterium] ventriosum group, Marvinbryantia, Colidextribacter and species [Clostridum] leptum. Bacteria that have an adverse effect (shortening of PFS or OS) on the response to treatment using immune checkpoint inhibitors are Rothia genus, Streptococus salivarius, Streptococus, Family XIII AD3011 group and Family XIII AD3011 group, s. uncultured bacterium. The composition of intestinal flora can be a predictive factor for immunotherapy in NSCLC patients. Specific bacteria can be positively or negatively associated with response to treatment, progression-free survival, and overall survival. They can be potentially used as predictive markers in NSCLC patients treated with immunotherapy.
Similar content being viewed by others
Introduction
The composition of the gut microbiome of patients with advanced non-small cell lung cancer (NSCLC) is currently being considered as one of the factors influencing the efficacy of treatment with immune checkpoint inhibitors (ICIs)1,2,3. Immune checkpoints PD-1 (Programmed Cell Death 1) and PD-L1 (Programmed Cell Death Ligand 1) are the targets for monoclonal antibodies. Immunotherapy is a breakthrough treatment for advanced NSCLC, resulting in prolonged patient survival and improved quality of life. However, some patients do not benefit from treatment. The primary resistance could occur as a progressive disease with short progression-free survival (PFS) and overall survival (OS). Secondary resistance against immunotherapy is characterized by initial benefit from immunotherapy followed by disease progression4,5,6. Mechanisms for the emergence of resistance to immunotherapy are currently being widely investigated. Potential reasons for the failure of ICI therapy include depletion of the immune system, including immune factors that extinguish its activity, genetic factors such as mutations in STK11 (Serine/Threonine Kinase 11), KRAS (KRAS Proto-Oncogene, GTPase), KEAP1 (Kelch Like ECH Associated Protein 1)genes, or epigenetic factors related to methylation or miRNA activity. Factors affecting the efficacy of immunotherapy in NSCLC patients also include the composition and diversity of the gut microbiome1. It has been indicated that the alpha and beta microbial diversity in a patient’s gut can affect the degree of clinical response to ICI’s. We aimed to evaluate the gut microbiome composition in NSCLC patients treated with first-line immunotherapy alone or in combination with chemoimmunotherapy. The possibility of assessing the microbiome’s composition has emerged through next-generation sequencing technology7,8,9. Using metataxonomic analysis, reads are assigned to the lowest possible taxonomic levels, allowing for the assessment of bacterial populations. However, due to limited sensitivity (as the analysis is based on two variable regions of the 16S rRNA gene, approximately 450 bp in length), this approach typically characterizes the microbiome at higher taxonomic levels, such as families, rather than more specific levels like genus or species7,8,9. Bacterial groups such as Ruminococcaceae, Lachnospiraceae or specific species, Akkermansia muciniphila, Faecalibacterium prausnitzii, which can be allies of immunotherapy, are pointed out. However, there are also bacteria which can hinder therapeutic results with ICI. Our study aims to show that bacteria residing in the gut can serve as biomarkers of the effectiveness of treatment, in this case, immunotherapy.
Materials and methods
Patients characteristic
For this prospective study, we enrolled patients with advanced NSCLC who were eligible for treatment with immunotherapy alone or in combination with chemotherapy. Experiments were performed following the Declaration of Helsinki. Informed consent was obtained from patients for the study. Patients whose material was available to receive before the implementation of immuno- or chemoimmunotherapy were eligible for the study. Patients’ characteristics are presented in Table 1. The study was performed with the approval (KE-0254/58/2019) of the Bioethics Committee at the Medical University of Lublin. Fifty-two patients treated in the first line were included in the study from May 2019 to August 2022. Squamous cell carcinoma was diagnosed in 21 (40%) patients, adenocarcinoma in 25 (48%), NOS in 4 (8%) and large cell carcinoma in 2 (4%) patients. The median age in the group was 66 years, with twenty-four patients below and 28 above the median. In the cohort, there were 27 (52%) men and 25 (48%) women. There were 3 (6%) patients in stage IIIB, while the remaining patients (n = 49, 94%) were in stage IV. All patients were negative for EGFR (Epidermal Growth Factor Receptor), ALK (Anaplastic Lymphoma Kinase), and ROS1 (ROS Proto-Oncogene 1) mutation and rearrangement, respectively. PD-L1 expression was assessed in routine diagnostic testing. In 44 (85%) patients, any PD-L1 expression was present, while in 8 (15%) patients, no PD-L1 expression was detected on tumor cells. In 33 patients, the percentage of tumor cells (TC) expressing PD-L1 was above 50%, and these patients were treated with ICI’s in monotherapy, while 19 had TC PD-L1(+) below 50%. They were treated with immunotherapy in combination with chemotherapy (Suplementary Table). Response to treatment was assessed according to RECIST 1.1 criteria. Six patients were treated with antibiotics up to four weeks before immunotherapy. The PFS was counted from the start of ICI. OS was calculated from the start of immunotherapy and follow-up to the endpoint of 31.01.2024. At this point, 10 (19%) patients remained alive.
Metataxonomics (16S metagenomics)
The study consisted of stool samples collected from patients who qualified for immunotherapy or chemoimmunotherapy. Samples were collected before treatment administration and immediately stored at −80oC until extraction. Twenty milligrams of stool was homogenized (FastPrep 24, MP Biomedicals) and treated with a cocktail of lysozyme (10 µg/ml, A&A Biotechnology) and lysostaphin (2000 U, Sigma-Aldrich) for 30 min in 37 °C. Extraction of total DNA was done by Maxwell RCS 48 (Promega) device according to RSC Tissue DNA Kit (Promega) protocol. The DNA was measured using Qubit 3.0 (Thermo Fisher Scientific) with High Sensitivity DNA Assay (Thermo Fisher Scientific). The sequencing library was prepared with 15 ng of total DNA according to 16S metagenomics protocol (Illumina). Quality check of libraries was performed by gel electrophoresis (Fragment Analyzer with dsDNA 935 Reagent Kit, Agilent). Normalization was done by fluorimeter (Qubit 3.0 with High Sensitivity Assay; Thermo Fisher Scientific). Pair-end sequencing (2 × 300 bp with V3 kit, Illumina) was performed on MiSeq (Illumina). The fastQ files were quality-checked with FastQC. All sample analyses were done using QIIME2 2023.9.2 platform10. For steps such as quality control, trimming, and filtering cutadatp plugin was used11,12,13. Additional reads trimming, denoising, chimera removal and eventually obtaining ASV table, have been made using dada2 plugin14. Sequence taxonomic annotation was perform by classify-sklearn plugin and SILVA 138 reference database15,16,17. For further statistical analysis, taxonomic results was collapsed to specific taxonomic level by collapse plugin, export to biom format and convert to tsv table by ‘biom18,19,20.
Sequencing data was uploaded to ENA repositories (BioProject ID: PRJNA1096150).
Statistical analysis
Statistical analyses of the reads were performed using Statistica 13.3 and MedCalc software on family and species taxonomic levels. These statistical software were also used to generate all the plots. The Mann-Whitney U test was used to analyze dichotomous variables. The Kruskal-Wallis test was performed to assess differences between patients with disease progression (PD), stable disease (SD) and partial response (PR). Survival curve analysis was performed using the Kaplan–Meier method. A p-value of less than 0.05 was considered statistically significant.
Results
Response to treatment
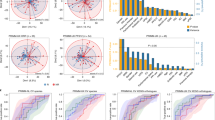
SD was observed in 16 (30%) of patients, PR was observed in 22 (40%) and PD was observed in 14 (30%) patients. Microbiome composition on the family level showed a higher percentage of Streptococcaceae in PD + SD patients than in patients with PR to ICI (p = 0.00071, Fig. 1a). The rate of Tannerellaceae is higher in PR patients compared to SD + PD (p = 0.043, Fig. 1b). If we regrouped patients with disease control as PR + SD, we found that patients with disease progression (PD) had a higher percentage of Micrococcaceae than the disease control group (p = 0.00049, Fig. 1c). We also found that the percentage abundance of Ruminococcaceae, Streptococcaceae, Tannerellaceae, Micrococcaceae, and Peptostreptococae was significantly different between PD, PR and SD group of patients as shown in Fig. 1d, e, f, g and h, respectively.
Performing deeper analysis on the species level, we found correlations between the content of bacteria and response to therapies in the first months of therapy efficacy evaluation (Fig. 2; Table 2). We noted a significantly higher rate of Ruminococcacea in patients with partial response and disease stabilization (PR + SD) compared to patients with disease progression (PD) (p = 0.012, Fig. 2a). We also noted a significantly higher percentage of G: Rothia and S: Streptococcus salivarius in patients who experienced disease progression compared to patients with partial response or disease stabilization (p = 0.00049, Fig. 2b and p = 0.018, Fig. 2c). For patients with PD or SD compared to patients with PR, we observed a higher percentage of G: Streptococcus (p = 0.0015, Fig. 2d) and G: Rothia (p = 0.0014, Fig. 2e) and a lower percentage of S: Alistipes sp. (p = 0.038, Fig. 2f). Figure 2g-m show the differences in content between the patients with PD, SD and PR groups.
PFS and OS analysis
The median PFS for the entire group was 5.5 months (95% CI: 3.23–6.57). The observations were complete in 46 patients, while six were censored. PFS of less than and above six months was observed in 31 and 21 patients, respectively. PFS of less than and above 12 months was observed in 39 and 13 patients, respectively.
The median OS was 9.9 months (95% CI: 6.6–27.9). We had complete data in 32 patients, while in 20, we had cut-off data. OS of less than and above six months was observed in 11 and 41 patients, respectively. OS of less than and above 12 months was observed in 31 and 21 patients, respectively.
We reported lower median percentages of Streptococcacea and Micrococcaceae with PFS over 6 months (p = 0.049, Fig. 3a.0037, Fig. 3b, respectively). PFS over 12 months was associated with a higher median percentage of Akkermansiaceae content (p = 0.046, Fig. 3c). Higher OS, over 6 or 12 months was associated with lower median Micrococcaceae content (p = 0.0079 and p = 0.0054, respectively, Fig. 3d).
Kaplan-Meier analyses showed that the presence of Micrococcaceae bacteria was associated with the risk of shortened PFS and OS (HR = 3.31, 95% CI: 1.71–6.42, p = 0.004, Fig. 4a and HR = 3.89, 95% CI: 1.79–8.46, p = 0.006, Fig. 4c respectively).
Median PFS and OS in patients with Micrococaceae present were 1.9 months and 5.2 months, respectively, and were lower compared to patients without identified bacteria from this family, which were 7.2 months and 27.9 months, respectively. Also, the presence of bacteria from the Gemellaceae family indicates the risk of shortened OS (HR = 2.54, 95% CI: 1.12–8.5.76, p = 0.026, Fig. 4d), and median OS was significantly lower compared to patients in whom the bacteria in this group was not identified (5.4 vs. 21.2 months). The presence of Oscillospirales uncultured is associated with a low risk of shortened PFS with HR = 0.48, 95% CI: 0.24–0.95, p = 0.035 (Fig. 4b). The median PFS of patients in the presence of these bacteria was 8.3 months, and in their absence was 4.9 months.
In species-level analysis, we observed in patients with a PFS of more than 6 months, a lower rate of G: Rothia (p = 0.0037, Fig. 5a) Streptococcus salivarius (p = 0.03, Fig. 5b) and G: Family XIII AD3011 group (p = 0.038, Fig. 5c) and a higher percentage of S: Alistipes sp. (0.049, Fig. 5d) compared to patients with a PFS of less than 6 months. In patients with a PFS of more than 12 months, we observed a lower rate of G: Family XIII AD3011 group, S: uncultured bacterium (p = 0.046, Fig. 5e), and a higher percentage of [Clostridium] leptum (p = 0.027, Fig. 5f) and G: Akkermansia (p = 0.048, Fig. 5g).
In patients with OS over 6 months, a higher percentage of F: Ruminococcacea (p = 0.0034, Fig. 5h), G: [Eubacterium] ventriosum group, p = 0.022, Fig. 5k), G: Marvinbryantia (p = 0.032, Fig. 5l), Colidextribacter (p = 0.023, Fig. 5m), [Clostridium] leptum (p = 0.036, Fig. 5n) and lower percentage of Rothia (p = 0.0054, Fig. 5i), Streptococcus salivarius (p = 0.021, Fig. 5j) compared to patients with OS less than 6 months.
In patients with OS over 12 months, we found a lower percentage of G: Rothia (p = 0.0054, Fig. 5o) and higher Marvinbryantia (p = 0.014, Fig. 5p) compared to patients with OS less than 12 months.
In Kaplan-Meier analyses, we showed that the presence of G: Rothia or G: Family XIII AD3011 group indicates the risk of shortening PFS with HR = 3.31, 95% CI: 1.71 to 6.42, p = 0.0004 (Fig. 6a) and HR = 2.48, 95% CI: 1.27 to 4.83, p = 0.0077 (Fig. 6c) respectively. In addition, the presence of Alistipes sp. or [Clostridium] leptum is associated with a low risk of shortened PFS with HR = 0.35, 95% CI: 0.17 to 0.72, p = 0.0042 (Fig. 6b), and HR = 0.43, 95% CI: 0.23 to 0.82, p = 0.01 (Fig. 6d) respectively.
Discussion
We analyzed the gut microbiome composition in patients with non-small cell lung cancer treated with first-line immuno- or chemoimmunotherapy. We first performed the analysis at the family taxonomic level, which provided an overall picture of which bacterial taxa would benefit from immunotherapy and which might indicate the success of ICI treatment.
The bacterial families Tannerellaceae, Oscillospirales genus: uncultured, Peptostreptococcae and Ruminococcaceae in our analyses were associated with longer PFS, OS and treatment response. As for Ruminococcacea, the study has shown that their number is significantly higher in patients with stable disease than in patients with partial response.
Analysis on a species level provided a broader perspective of the relationship between the bacteria present in the gut of NSCLC patients and the efficacy of ICI. We linked the Ruminococcaceae family with SD/PR and PFS longer than 6 months. A study by Gopalakrishnan et al. reported that Ruminococcaceae were enriched in response to anti-PD-1 melanoma treatment21. In addition to melanoma, patients suffering from NSCLC, and kidney cancer who respond to ICI in the gut also have more Ruminoccocus compared to non-responders21,22,23,24,25,26,27. Jin et al. found Ruminococcus_unclassified enriched in non-responder patients with advanced NSCLC28. Our study presented the Ruminococcaceae family as occurring in high percentages of patients with stable disease. It is known that some patients treated with immunotherapy experience secondary resistance to treatment despite early responses to immunotherapy, and perhaps this group of microorganisms should be looked at in this aspect. This bacterium potential can predict the emergence of acquired resistance to ICI. However, this is only a hypothesis that needs to be confirmed in studies. This is particularly important because this bacterium is primarily associated with patients who respond to treatment; however, there have been studies linking representatives of this group to non-response in NSCLC treatment28,29. Gopalakrishnan et al. indicate that Ruminococcaceae in melanoma patients are highly-represented in patients responding to anti-PD-1 therapy. They also state that Rothia is associated with a favourable course of treatment21. Takabe et al. showed that Rothia in the tissue of NSCLC was associated with worse survival. They found that Rothia was the only bacterium whose detection in the microbiome of NSCLC tumors was significantly associated with shorter patient survival30. Naddaf et al. and Zhao et al. indicated that Rothia dentocariosa levels were increased in the stool of patients with lung and hematologic cancers, as well as those with a history of Lynch syndrome and cancer. This increase correlates with poorer treatment efficacy and shorter progression-free survival31,32. NSCLC tissue with Rothia was associated with worse survival and favorable oncological characteristics such as less cell proliferation and favorable tumor immune microenvironment. The authors speculate that Rothiain NSCLC tissue is associated with mortality unrelated to oncological characteristics30.
Our study on the family Streptococcaceae and genus Streptococcus level found that they are more abundant in patients with stable or progressive disease than those with partial response. Further analyses revealed that a high level of Streptococcus salivarius can be associated with a lack of response to ICI, PFS, and OS of less than 6 months. Dora et al. studied 62 patients with advanced NSCLC treated with ICI. Our group included 52 patients treated with first-line either immunotherapy or chemoimmunotherapy. In the Dora et al. study, 30 patients received ICI in the first line, and 32 patients received ICI in the following line. Dora et al. also indicated that patients with progression-free survival of less than 6 months positively correlated with Streptococcus salivarius33. A study of the composition of the microbiome was conducted by Peiffer et al. in patients with metastatic castration-resistant prostate cancer who had progressed after enzalutamide and had started anti-PD-1 treatment34. They showed an increased level of S. salivarius in fecal samples in responders to treatment. Moreover, they suggest that the association between fecal microbiota and treatment response to immunotherapy may be unique to cancer type or previous treatment history34.
The following groups of bacteria found unfavorable for ICI response in our study were the Family XIII AD3011 group and the Family XIII AD3011 group s. uncultured bacterium. The first one indicates short PFS (less than 6 months). Its presence increases the risk of shortened PFS and OS less than 12 months. The second indicates a PFS of less than 12 months. Lou et al. suggest that the Family XIII AD3011 group in basal cell carcinoma is a promoter of the occurrence of this type of carcinoma. However, whether these groups can promote the growth of carcinoma cells by influencing the migration and transformation of other cells requires further targeted research for validation35. Xie et al. found that the genus Family XIII AD3011 group was negatively associated with the risk of liver cancer36. In the study, Park et al. showed in stool sample study that the relative abundance of the genus Family XIII AD3011 group was lower in the colorectal patients than in the control group (70 CRC subjects and 158 control subjects)37.
In our study, the beneficial gut bacteria identified were species of Alistipes sp. Their presence was associated with prolonged progression-free survival (PFS) and a higher likelihood of achieving a PFS of more than six months. Furthermore, these bacteria were linked to a better response to treatment than patients with a PFS of less than six months and those with no response to treatment or disease stabilization. Other studies suggest that Alistipes sp. species may predict response to ICI and longer survival times2,22,33,38. Dora lists Alistipes shahii and Alistipes finegoldii as being associated with PFS beyond 6 months in patients with NSCLC treated with anti-PD-1. Patients with liver cancer who responded to anti-PD-1 therapy showed a higher abundance of Alistipes sp. Marseille-P5997, and this was associated with longer PFS and overall survival in patients with inoperable liver and biliary tract cancers39.
We also observed the genus [Eubacterium] ventriosum group as a positive predictor of ICI effectiveness. Its higher percentage is seen in patients with OS above compared to those below 6 months, and its low percentage is associated with non-response to treatment and disease progression. In Hokazaki study, the patients without cachexia were enriched with beneficial bacteria, including Eubacterium40. In 113 Japanese patients with advanced NSCLC treated with ICIs (79 in first-line monotherapy and 34 in second or later-line treatment), Eubacterium ventriosum is associated with longer PFS and OS40. In addition, Eubacterium ventriosum was presented by Zhang et al. as enriched in the high immune-related adverse events (irAE) group compared to the low-grade irAE group in patients with esophageal cancer treated with ICI41. This is an interesting observation because irAEs were associated with better tumor response to treatment in patients with advanced NSCLC treated with ICIs42.
The Marvinbrantia genus also favored patient response and more prolonged OS of more than 6 and 12 months, compared to groups with disease stabilization or progression and OS of less than 6/12 months, respectively. Also, Colidextribacter appeared as a genus associated with OS that was higher than 6 months. Newsome et al. also indicate that the taxa associated with response to immunotherapy in NSCLC patients at stage IIIA-IV include Colidextribacter and Marvinbryantia43. Their group had a size of 65 III/IV NSCLC, 37 of whom had been previously treated with chemotherapy. It is important to remember that pre-immuno treatment can influence the microbiome’s composition. Therefore, our study focused on patients treated only in the first-line ICI alone or in combination with chemotherapy. Research is also thought to be needed to assess potential microbiological predictors of response to chemotherapy, immunotherapy or combination treatment.
Akkermansia muciniphila (A. muciniphila), described in the microbiome literature related to the efficacy of ICI, can significantly inhibit carcinogenesis and improve antitumor activity44. Fan et al. indicate that A. muciniphila is involved in host immune response and maintenance of intestinal barrier integrity44. In our study, we show that a higher percentage of the Akkermansia genus in the gut microbiome, with A. muciniphila as the leading representative, is associated with longer progression-free survival (PFS), precisely more than 12 months. A. muciniphila is widely reported in the literature as a favorable microorganism for immunotherapy. Nevertheless, the mechanisms involved in its action and beneficial effects on the efficacy of immunotherapy are not fully understood22,45,46. Derosa et al. indicate that the amount of A. muciniphila prior to treatment implementation significantly predicts response to immunotherapy in patients with advanced NSCLC, even better than PD-L1 expression. In addition, they indicate that the relative amount of A. muciniphila is a better predictor of response expressed in OS length than the presence or absence of these bacteria45. We have also shown that the presence of the Clostridium leptum group ([Clostridium] leptum) is associated with prolonged PFS, with a higher percentage of this bacterium indicating OS over six months. Gui et al. indicate that Faecalibacterium prausnitzii and Clostridium leptum, belongers of Clostridium leptum group, showed significantly decreased NSCLC compared to healthy individuals47. Clostridium spp. is a broad genus, and this group includes Clostridium cluster IV (Clostridium leptum group), and the classification of Clostridium spp. is constantly changing due to more and more metagenomic data and 16S rRNA sequencing48.
F. prausnitzii has probiotic immunomodulatory potential, and patients receiving ICI who developed colitis had lower numbers of this microorganism, which enhances the tumor-suppressive effects of immune checkpoint blockade49. Faecalibacterium prausnitzii, has been linked to favorable outcomes in melanoma, lung cancer and renal cell carcinoma patients treated with immunotherapy50. In melanoma, patients who responded to ipilimumab plus nivolumab had increased gut bacterial species, including F. prausnitzii51. The differences in research on the microbiome of oncology patients, as indicated by Zhang et al. or Zeriouh et al. In their reviews, the authors indicate bacteria from the families Ruminococcaceae, Akkermasiacea, Bifidobacterium and Barnesiellaceae, which are considered beneficial or not for oncological patients. This may be due to various factors, such as the type of cancer and the type of immunotherapy used, the size of the group, treatment before the implementation of immunotherapy, or the method used to assess the microbiome composition2,29,34. It should also be mentioned that NGS and metagenomic analysis are very good tools for screening and providing more and more data. The resulting data introduce taxonomic reshuffling in microbiology. Therefore, tests identifying individual bacteria, selected through metagenomics, can be based on other methods, such as targeting, for example, qPCR, which will evaluate a narrower panel of selected bacteria. It is worth to mention that this is not a study of the functionality of the bacteria in question. The value of this study is because extensive screening with NGS is the starting point for selecting individual bacteria at particular taxonomic levels, which in the future can be used in daily clinical practice as prognostic-predictive factors.
Conclusion
We conclude that the beneficial bacteria for immunotherapy of NSCLC patients included F: Ruminococcaceae, S: Alistipes sp., G: [Eubacterium] ventriosum group, G: Marvinbryantia, G: Akkermansia, G: Colidextribacter, S: [Clostridum] leptum. Conversely, bacteria that adversely affect the response to ICI, shortening progression-free survival (PFS) or overall survival (OS), include G: Rothia, S: Streptococus salivarius, G: Streptococus, G: Family XIII AD3011 group, G: Family XIII AD3011 group, s. uncultured bacterium. Further research in this area should be conducted to provide more information on how the microbiome and its bacterial components affect the effectiveness of immunotherapy in NSCLC patients treated with first-line ICIs. This could potentially allow for selecting individual bacterial strains that serve as clinically useful markers of treatment response. Further study is needed to see if the same correlations occur in the immunotherapy group alone as in the group of patients treated with concurrent chemotherapy.Disclosure statement
Data availability
The data that support our findings of this study are available at http://www.ncbi.nlm.nih.gov/bioproject/1096150.
References
Shah, H. & Ng, T. L. A narrative review from gut to lungs: non-small cell lung cancer and the gastrointestinal microbiome. Transl Lung Cancer Res. 12, 909–926. https://doi.org/10.21037/tlcr-22-595 (2023).
Zhang, M., Liu, J. & Xia, Q. Role of gut microbiome in cancer immunotherapy: from predictive biomarker to therapeutic target. Exp. Hematol. Oncol. 12, 84. https://doi.org/10.1186/s40164-023-00442-x (2023).
Verschueren, M. V. et al. The association between gut microbiome affecting concomitant medication and the effectiveness of immunotherapy in patients with stage IV NSCLC. Sci. Rep. 11, 23331. https://doi.org/10.1038/s41598-021-02598-0 (2021).
Gandhi, L. et al. Pembrolizumab plus Chemotherapy in Metastatic non–small-cell Lung Cancer. N Engl. J. Med. 378, 2078–2092. https://doi.org/10.1056/NEJMoa1801005 (2018).
Topalian, S. L. et al. Five-year survival and correlates among patients with Advanced Melanoma, Renal Cell Carcinoma, or Non-small Cell Lung Cancer treated with Nivolumab. JAMA Oncol. 5, 1411–1420. https://doi.org/10.1001/jamaoncol.2019.2187 (2019).
West, H. et al. Atezolizumab in combination with carboplatin plus nab-paclitaxel chemotherapy compared with chemotherapy alone as first-line treatment for metastatic non-squamous non-small-cell lung cancer (IMpower130): a multicentre, randomised, open-label, phase 3 trial. Lancet Oncol. 20, 924–937. https://doi.org/10.1016/S1470-2045(19)30167-6 (2019).
Zhang, J. et al. Application of Microbiome in Forensics. Genomics Proteom. Bioinf. 21, 97–107. https://doi.org/10.1016/j.gpb.2022.07.007 (2023).
Wensel, C. R., Pluznick, J. L., Salzberg, S. L. & Sears, C. L. Next-generation sequencing: insights to advance clinical investigations of the microbiome. J. Clin. Invest. 132, e154944. https://doi.org/10.1172/JCI154944
Khachatryan, L. et al. Taxonomic classification and abundance estimation using 16S and WGS—A comparison using controlled reference samples. Forensic Sci. Int. Genet. 46, 102257. https://doi.org/10.1016/j.fsigen.2020.102257 (2020).
Bolyen, E. et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. https://doi.org/10.1038/s41587-019-0209-9 (2019).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. https://doi.org/10.14806/ej.17.1.200 (2011).
McDonald, D. et al. The Biological Observation Matrix (BIOM) format or: how I learned to stop worrying and love the ome-ome. GigaScience 1, 7 (2012). https://doi.org/10.1186/2047-217X-1-7
Pedregosa, F. et al. Scikit-learn: Machine Learning in Python. Mach Learn PYTHON (2011).
Callahan, B. J. et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods. 13, 581–583. https://doi.org/10.1038/nmeth.3869 (2016).
Pruesse, E. et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35, 7188–7196. https://doi.org/10.1093/nar/gkm864 (2007).
Quast, C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–596. https://doi.org/10.1093/nar/gks1219 (2013).
Robeson, M. S. et al. RESCRIPt: reproducible sequence taxonomy reference database management. PLoS Comput. Biol. 17, e1009581. https://doi.org/10.1371/journal.pcbi.1009581 (2021).
Bokulich, N. A. et al. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6, 90. https://doi.org/10.1186/s40168-018-0470-z (2018).
Rognes, T. et al. VSEARCH: a versatile open source tool for metagenomics. PeerJ 4, e2584. https://doi.org/10.7717/peerj.2584 (2016).
Data Structures for Statistical Computing in Python - SciPy Proceedings. (2010). https://proceedings.scipy.org/articles/Majora-92bf1922-00a. Accessed 30 Oct 2024.
Gopalakrishnan, V. et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359, 97–103. https://doi.org/10.1126/science.aan4236 (2018).
Routy, B. et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97. https://doi.org/10.1126/science.aan3706 (2018).
Tanoue, T. et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature 565, 600–605. https://doi.org/10.1038/s41586-019-0878-z (2019).
Davar, D. et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science 371, 595–602. https://doi.org/10.1126/science.abf3363 (2021).
Smith, M. et al. Gut microbiome correlates of response and toxicity following anti-CD19 CAR T cell therapy. Nat. Med. 28, 713–723. https://doi.org/10.1038/s41591-022-01702-9 (2022).
Zhang, H. & Xu, Z. Gut–lung axis: role of the gut microbiota in non-small cell lung cancer immunotherapy. Front. Oncol. 13 https://doi.org/10.3389/fonc.2023.1257515 (2023).
Hakozaki, T. et al. The Gut Microbiome Associates with Immune Checkpoint Inhibition outcomes in patients with Advanced Non-small Cell Lung Cancer. Cancer Immunol. Res. 8, 1243–1250. https://doi.org/10.1158/2326-6066.CIR-20-0196 (2020).
Jin, Y. et al. The diversity of gut microbiome is Associated with favorable responses to anti–programmed death 1 immunotherapy in Chinese patients with NSCLC. J. Thorac. Oncol. 14, 1378–1389. https://doi.org/10.1016/j.jtho.2019.04.007 (2019).
Zeriouh, M. et al. Checkpoint inhibitor responses can be regulated by the gut microbiota - A systematic review. Neoplasia N Y N. 43, 100923. https://doi.org/10.1016/j.neo.2023.100923 (2023).
Takabe, Y. J. et al. Rothia in Nonsmall Cell Lung Cancer is Associated with worse survival. J. Surg. Res. 296, 106–114. https://doi.org/10.1016/j.jss.2023.12.026 (2024).
Naddaf, R. et al. Gut microbial signatures are associated with Lynch syndrome (LS) and cancer history in druze communities in Israel. Sci. Rep. 13, 20677. https://doi.org/10.1038/s41598-023-47723-3 (2023).
Zhao, Z. et al. Metagenome association study of the gut microbiome revealed biomarkers linked to chemotherapy outcomes in locally advanced and advanced lung cancer. Thorac. Cancer. 12, 66–78. https://doi.org/10.1111/1759-7714.13711 (2021).
Dora, D. et al. Non-small cell lung cancer patients treated with Anti-PD1 immunotherapy show distinct microbial signatures and metabolic pathways according to progression-free survival and PD-L1 status. Oncoimmunology 12, 2204746. https://doi.org/10.1080/2162402X.2023.2204746.
Peiffer, L. B. et al. Composition of gastrointestinal microbiota in association with treatment response in individuals with metastatic castrate resistant prostate cancer progressing on enzalutamide and initiating treatment with anti-PD-1 (pembrolizumab). Neoplasia N Y N. 32, 100822. https://doi.org/10.1016/j.neo.2022.100822 (2022).
Lou, J. et al. Causal relationship between the gut microbiome and basal cell carcinoma, melanoma skin cancer, ease of skin tanning: evidence from three two-sample mendelian randomisation studies. Front. Immunol. 15 https://doi.org/10.3389/fimmu.2024.1279680 (2024).
Xie, N. et al. Association between Gut Microbiota and Digestive System cancers: a bidirectional two-sample mendelian randomization study. Nutrients 15, 2937. https://doi.org/10.3390/nu15132937 (2023).
Park, J. et al. Fecal microbiota and gut microbe-derived extracellular vesicles in Colorectal Cancer. Front. Oncol. 11, 650026. https://doi.org/10.3389/fonc.2021.650026 (2021).
Duttagupta, S., Hakozaki, T., Routy, B. & Messaoudene, M. The gut microbiome from a biomarker to a Novel Therapeutic Strategy for Immunotherapy Response in patients with Lung Cancer. Curr. Oncol. 30, 9406–9427. https://doi.org/10.3390/curroncol30110681 (2023).
Mao, Z. et al. First-line immuneâ€based combination therapies for advanced nonâ€small cell lung cancer: a bayesian network metaâ€analysis. Cancer Med. 10, 9139–9155. https://doi.org/10.1002/cam4.4405 (2021).
Hakozaki, T. et al. Cancer Cachexia among patients with Advanced Non-small-cell Lung Cancer on Immunotherapy: an observational study with exploratory gut microbiota analysis. Cancers 14, 5405. https://doi.org/10.3390/cancers14215405 (2022).
Zhang, Y. et al. Correlation of the gut microbiome and immune-related adverse events in gastrointestinal cancer patients treated with immune checkpoint inhibitors. Front. Cell. Infect. Microbiol. 13, 1099063. https://doi.org/10.3389/fcimb.2023.1099063 (2023).
Sung, M. et al. Correlation of immune-related adverse events and response from immune checkpoint inhibitors in patients with advanced non-small cell lung cancer. J. Thorac. Dis. 12, 2706–2712. https://doi.org/10.21037/jtd.2020.04.30 (2020).
Newsome, R. C. et al. Interaction of bacterial genera associated with therapeutic response to immune checkpoint PD-1 blockade in a United States cohort. Genome Med. 14, 35. https://doi.org/10.1186/s13073-022-01037-7 (2022).
Fan, S. et al. Akkermansia muciniphila: a potential booster to improve the effectiveness of cancer immunotherapy. J. Cancer Res. Clin. Oncol. 149, 13477–13494. https://doi.org/10.1007/s00432-023-05199-8 (2023).
Derosa, L. et al. Intestinal Akkermansia muciniphila predicts clinical response to PD-1 blockade in patients with advanced non-small-cell lung cancer. Nat. Med. 28, 315–324. https://doi.org/10.1038/s41591-021-01655-5 (2022).
Routy, B. et al. The gut microbiota influences anticancer immunosurveillance and general health. Nat. Rev. Clin. Oncol. 15, 382–396. https://doi.org/10.1038/s41571-018-0006-2 (2018).
Gui, Q. et al. The association between gut butyrate-producing bacteria and nonâ€smallâ€cell lung cancer. J. Clin. Lab. Anal. 34, e23318. https://doi.org/10.1002/jcla.23318 (2020).
Grenda, T. et al. Probiotic potential of Clostridium spp.—Advantages and doubts. Curr. Issues Mol. Biol. 44, 3118–3130. https://doi.org/10.3390/cimb44070215 (2022).
Gao, Y. et al. Faecalibacterium prausnitzii abrogates intestinal toxicity and promotes Tumor Immunity to increase the efficacy of dual CTLA4 and PD-1 checkpoint blockade. Cancer Res. 83, 3710–3725. https://doi.org/10.1158/0008-5472.CAN-23-0605 (2023).
Chang, J. W. C. et al. Gut microbiota and clinical response to immune checkpoint inhibitor therapy in patients with advanced cancer. Biomed. J. 100698. https://doi.org/10.1016/j.bj.2024.100698 (2024).
Frankel, A. E. et al. Metagenomic shotgun sequencing and unbiased metabolomic profiling identify specific human gut microbiota and metabolites Associated with Immune Checkpoint Therapy Efficacy in Melanoma patients. Neoplasia N Y N. 19, 848–855. https://doi.org/10.1016/j.neo.2017.08.004 (2017).
Author information
Authors and Affiliations
Contributions
A.G., P.K., E.I., T.J.: designed the study. I.Ch. M. F., B. KK, R.K.: collected and interpreted the data. A.B., K.B., A.G., M.Sz., I.Ch, A.RK., : analyzed and interpreted the data. A.G. E.I., I.Ch., P.K. prepared the manuscript. J.M., P.K. supervision and critical revision. All authors approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Grenda, A., Iwan, E., Kuźnar-Kamińska, B. et al. Gut microbial predictors of first-line immunotherapy efficacy in advanced NSCLC patients. Sci Rep 15, 6139 (2025). https://doi.org/10.1038/s41598-025-89406-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-89406-1