Abstract
Alzheimer’s disease (AD) is the most common form of dementia in the elderly, affecting cognitive, intellectual, and motor functions. Different hypotheses explain AD’s mechanism, such as the amyloidogenic hypothesis. Moreover, this disease is multifactorial, and several studies have shown that gut dysbiosis and oxidative stress influence its pathogenesis. Knowing that kefir is a probiotic used in therapies to restore dysbiosis and that the bioactive peptides present in it have antioxidant properties, we explored its biotechnological potential as a source of molecules capable of modulating the amyloidogenic pathway and reducing oxidative stress, contributing to the treatment of AD. For that, we used Drosophila melanogaster model for AD (AD-like flies). Identification of bioactive peptides in the kefir sample was made by proteomic and peptidomic analyses, followed by in vitro evaluation of antioxidant and acetylcholinesterase inhibition potential. Flies were treated and their motor performance, brain morphology, and oxidative stress evaluated. Finally, we performed molecular docking between the peptides found and the main pathology-related proteins in the flies. The results showed that the fraction with the higher peptide concentration was positive for the parameters evaluated. In conclusion, these results revealed these kefir peptide-rich fractions have therapeutic potential for AD.
Similar content being viewed by others
Introduction
Alzheimer’s disease (AD) is a multifactorial progressive pathology and the most common form of dementia1. It compromises distinct cognitive abilities, such as learning and memory, motor performance, and visual-spatial function2. In 2019, AD caused 1.5 million deaths, and by 2050 global cases are estimated to reach 175.9 million3. It is neuropathological characterized by lesions and atrophy in brain regions and the main hypothesis for its development is the aggregation of beta-amyloid peptides4. Those 42 amino acid peptides are generated through the amyloidogenic pathway, in which the amyloid precursor protein (APP) is cleaved by a β-secretase (BACE) followed by a γ-secretase4,5,6.
Recent studies indicate that AD’s pathology is directly correlated to dysbiosis in the gut-brain axis, inducing inflammatory and oxidative responses, while also influencing β-amyloid aggregation7,8. In this context, probiotics administration have demonstrated a positive effect in AD animal models and patients9,10. Among those, kefir is an especially promising probiotic. It is constituted by symbiotic bacteria and yeasts, which species and ratio can vary according to geographical origin and substrate—most commonly cow milk11,12. In our previous studies, kefir in natura and its metabolites decreased amyloid aggregation and improved the motor capacity of a Drosophila melanogaster model for AD (AD-like flies)13. Kefir’s bioactive peptides, however, are still to be explored against AD.
D. melanogaster has been extensively used to explore AD’s neurobiology and possible treatments14,15. It has favorable genetic characteristics and toolboxes, complex behavior, and a simplified nervous system and gut-brain axis—although conserved in relation to higher organisms. It is a unique model for exploring probiotic treatments to attenuate Alzheimer’s symptoms, especially targeting the amyloidogenic pathway16,17,18,19.
In order to provide a better understanding of the effect of Kefir’s bioactive peptides in AD’s pathology, we treated the AD-like flies with kefir and evaluated behavior and oxidative stress markers, as well molecular docking and in vitro approach.
Results
Kefir proteome and peptidome
Protein fractions (> and < 10 kDa) were analyzed by proteomic approaches. When the peptides were analyzed using the database “milk AND bovine”, a similar profile of peptides from milk proteins was found in both fractions, except for α-S1-casein, which was found only in the > 10 kDa fraction (Fig. 1a,b).
Proteome and peptidome of the kefir sample. (a) Proteomic analysis of the > 10 kDa fraction digested with trypsin and relationship of the proteins. (b) Proteomic analysis of the < 10 kDa fraction digested with trypsin and relationship of the proteins. (c) Peptidomic analysis of the < 10 kDa non-digested fraction and the quantity of peptides found that are derived from milk. Parts of the figure were drawn by using pictures from Servier Medical Art (https://smart.servier.com/).
The peptidome analysis of < 10 kDa fraction was performed by mass spectrometry without trypsin digestion. Peptides derived from α-S1-casein, β-casein and only one from β-lactoglobulin were found in the main milk protein (Fig. 1c). Peptides were also found from the Lactobacillus and Acetobacter databases. However, in this work, we focused only on peptides encrypted in milk proteins.
In silico prediction of bioactivities
The prediction of the physicochemical parameters and bioactivities of peptides from the “milk AND bovine” database and de novo analysis showed a large number of peptides with antioxidant, immunomodulatory, and ACE inhibitory activity, among others (Supplementary Table S1).
In vitro analysis of kefir fraction
As a first step to analyze the diversity of kefir water-soluble fractions (WSF), the fractions were tested for their acetylcholinesterase inhibition and antioxidant capacity in vitro.
Ascorbic acid was used as a positive control for the FRAP test20; as a pure compound, it demonstrated the highest antioxidant capacity. In this in vitro antioxidant assay, all tested fractions, i.e. the WSF and peptides (> 10 kDa and < 10 kDa), demonstrated antioxidant capacity in terms of converting Fe3+ into Fe2+ (Fig. 2a). With improvements in the filtering and purification processes, the antioxidant capacity increased, with the < 10 kDa/peptidic fraction showing the best performance (p < 0.5) (Fig. 2a).
In vitro analysis of the effects of kefir fractions. (a) Total antioxidant capacity by the FRAP method of Fe3+ reduction analysis. The < 10 kDa fraction shows higher antioxidant activity compared to the others (*p < 0.5), but not as high as ascorbic acid used as the control. (b) Acetylcholinesterase enzyme inhibition capacity. Among the kefir fractions, the < 10 kDa fraction stands out as having the highest inhibition capacity (*indicates p < 0.5).
In the in vitro acetylcholinesterase inhibition assay, the fractions showed a similar pattern, with the peptidic fraction < 10 kDa showing the highest inhibitory activity (p < 0.5) (Fig. 2b). However, the WSF fraction did not inhibit acetylcholinesterase.
Drosophila melanogaster AD model
After observing the promising activity of the fractions in preliminary in vitro studies, we used the fractions in a D. melanogaster model of Alzheimer’s disease (AD-like). This model is based on the expression of human BACE and APP, which mimics the amyloidogenic pathway in the fruit fly brain.
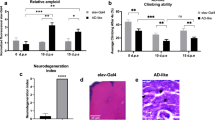
To ensure that the AD model is suitable for the evaluation of the fractions, we evaluated motor ability, performed β-amyloid quantification, and completed a histopathological analysis. For these assays, elav-Gal4/+ flies were used as the control.
To evaluate a decline in motor reflex behavior, linked to neurodegeneration, a negative geotaxis assay was used. As expected, 10-to 13-day-old flies presented impaired motor performance when compared to the control genotype (p < 0.01) and to 5- to 8-day-old flies (p < 0.001) (Fig. 3a).
Validation of the Alzheimer’s disease model. (a) Climbing ability. AD-like flies show reduced motor ability compared to the control genotype at 10–13 days post eclosion (n = 90 in each genotype). (b) Quantification of amyloid by the Thioflavin T method. AD-like flies showed a higher amount of amyloid compared to the control at 10–13 days post eclosion (n = 30 for each genotype). (c) Neurodegeneration index of elav and AD-like flies based on the histopathological analysis, focusing on vacuolar lesions. Indices range from 0 to 5 with 0 indicating no lesions and 5 indicating a neurodegenerative phenotype. Data are presented as mean ± SEM, and significance values are represented as **p < 0.0001, *p < 0.001, **p < 0.01. Representative histopathological images of the elav (d) and AD-like (e) genotypes.
Using the amyloid binding capacity of Thioflavin T21, we quantified the relative amyloid levels in 10- to 13-day-old flies (n = 30). AD-like flies had a higher level of β-amyloid than control flies (p < 0.01, Fig. 3b).
Finally, the brains of 10-to 13-day-old flies underwent a histopathological analysis to evaluate the neurodegeneration index, ranging from 0 to 5, as observed in elav-Gal4 (Fig. 3d) and AD-like flies (Fig. 3e), in accordance with previous studies13,22. These results point to a greater degree of neurodegeneration in AD-like flies compared to control flies (p < 0.5, Fig. 3c).
The peptide fraction from kefir can modulate the AD model
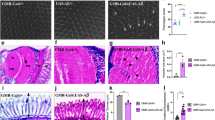
Our previous work demonstrated a positive effect of kefir in natura and its compounds in this AD-like model13. In the RING assay, treated flies showed an improvement in climbing ability starting at 10 days after treatment (and therefore 10–13 days post eclosion) when treated with the WSF (p < 0.5) and < 10 kDa (p < 0.01) fractions at 0.25 mg/mL (Fig. 4a), compared to untreated flies. Treatment with fractions at 0.5 mg/mL did not show any improvements in climbing (Fig. 4b). In order to analyze the effect of the fractions in other parameters, follow up assays were done with flies at 10–13 days post eclosion, treated with < 10 kDa and < 10 kDa fractions at 0.25 and 0.5 mg/mL.
In vivo effects of kefir fractions. (a) The climbing ability of AD-like flies after 5 and 10 days of treatment with the fractions at a concentration 0.25 mg/mL, flies treated with WSF and the < 10 kDa fraction showed an improvement in motor performance (n = 90). (b) The climbing ability of AD-like flies after 5 and 10 days of treatment with the fractions at a concentration 0.5 mg/mL (n = 90). (c) Quantification of amyloid content by the Thioflavin T assay. Flies treated with all fractions at both concentrations showed a reduction in the amyloid content compared to the control (untreated) at 10 days of treatment (n = 30 for each treatment). (d) Index of neurodegeneration based on the histopathological analysis according to vacuolar lesions (n = 10 at 10 days of treatment). The results show a decrease in the index in all treated flies compared to the control. (e) Fe3+ reduction capacity by the FRAP method. Flies treated with the fractions show reduced antioxidant activity (n = 30 and 10 days of treatment). (f) Acetylcholinesterase activity. Only flies treated with the < 10 kDa fraction at 0.25 mg/mL demonstrated decreased acetylcholinesterase activity. Data are presented as mean + SEM. Statistically significant differences are indicated by *p < 0.5, **p < 0.1, ***p < 0.01 and ****p < 0.001.
The t-test analysis showed that both fractions demonstrated a significant effect when administrated at 0.25 and 0.5 mg/mL. Flies treated with these samples showed a lower amyloid content (Fig. 4c) compared to control, neurodegeneration index (Fig. 4d) and antioxidant activity (Fig. 4e) when compared with untreated flies.
Exceptionally, in the anti-acetylcholinesterase assay (Fig. 4f), only flies treated with the < 10 kDa fraction (at 0.25 mg/mL) displayed a decrease in acetylcholinesterase activity when compared to control.
Molecular docking
The < 10 kDa peptide fraction from kefir had antioxidant activity, improved climbing ability, decreased beta-amyloid levels and inhibited acetylcholinesterase activity. Molecular docking was performed to verify the peptides present in the fraction identified in the peptidomic assay in terms of BACE, amyloid fibrils and acetylcholinesterase.
Nine peptides were used for the prediction of 3D structure and docking. All peptides contributed atomic contact energy to the global energy (ACE) below − 6 in the docking prediction for BACE, Aβ(1–42) fibrils and human acetylcholinesterase (Table 1). The best model for docking (the peptide with the lowest ACE value for each protein) is shown in Fig. 5.
Molecular docking analysis of the VPPFLQPEV and VYPFPGPIPN peptides and their interaction with BACE1, Aβ and acetylcholinesterase. (a) Prediction of the interaction of the VPPFLQPEV peptide with the enzyme BACE1. (b) Zoomed in image of panel (a). (c,d) Prediction of the interaction of the VYPFPGPIPN peptide with β-amyloid plaques between the first and second β-strand regions. (e) Hydrophobic region of interaction of the VYPFPGPIPN peptide with β-amyloid plaque. (f) Prediction of the interaction of the VYPFPGPIPN peptide in the peripheral anionic site of the acetylcholinesterase enzyme. (g) Zoomed in image of panel (f).
The peptide VPPFLQPEV was predicted as the best ligand for BACE. This peptide binds to an important flap that controls enzyme activity by partially covering the substrate-binding cleft localized between the N- and C-terminus lobes (Fig. 5a,b).
The best predicted ligand for Aβ(1–42) fibrils was the peptide VYPFPGPIPN. It could bind between the first and second β-strand regions (Fig. 5c,d). This is a hydrophobic region, and the binding of the peptide in this region may make it more hydrophobic (Fig. 5e).
The peptide VYPFPGPIPN was also predicted as the best ligand for human AChE. This enzyme has two binding sites: one is a peripheral anionic site (PAS) located at the entrance of the active gorge and the other is a catalytically active site (CAS) located at the base of the active site gorge. The peptide bound to PAS (Fig. 5f,g).
Discussion
Kefir is a broadly used probiotic, characterized as a symbiotic agglomerate of bacteria and yeasts. It is popularly used as a complementary treatment in many diseases23,24,25,26. Throughout the milk fermentation process, kefir microorganisms present a unique protein metabolic process, resulting in bioactive molecules27 among these peptides. The composition of kefir grains varies according to geographic location and fermentation parameters11,28,29 and is thought to contribute to variations in its metabolites and peptides. Even though there is variability, some core bacteria and yeasts remain consistent across samples, which allows for the comparison of different studies12. Our group has already characterized the effects of raw kefir and its organic fractions—as well as its metabolome—in improving AD-like flies13. This present work is a complementary effort, investigating the effects of kefir peptides in AD-like flies, along with a proteomic and peptidomic characterization. Additionally, this is the first study, to our knowledge, to characterize both peptides and metabolites from a single kefir grain sample.
In the present work, we focused on the peptidome of the kefir and in the intact bioactive peptides. The fermentation-generated peptides were identified through a proteomic analysis. Similar results have been described by other research groups30,31, which supports the compositional consistency of kefir despite some variations across samples.
As we used cell-free protein to produce the proteomic profile, the peptidases found using databases from Lactobacillus and Acetobacter could be from cells disrupted (intracellular) by fermentation or by a novel secretory protein degradation system32. Further assays are needed to resolve this question. In previous work, only these two bacteria genera were found in our kefir sample13.
Our in silico analysis indicates that the major part of the peptides generated from our kefir sample during fermentation display several activities, the characteristics of which have been described in other milk-derived peptides33,34. Here, we focused on putative acetylcholinesterase inhibition and antioxidant properties as well BACE inhibition and putative amyloid fibril binding.
These characteristics are especially interesting for AD. Since AD is a multifactorial disease35, researchers have turned their attention to developing multi-target drugs to inhibit the myriad factors involved in AD, including protein misfolding and associated Aβ aggregation, t aggregation, metal dyshomeostasis, oxidative stress and a decrease in AChE levels. Even though the proposed model is based on the amyloidogenic hypothesis, we should expect alterations in other metabolic processes. Previous studies have shown a relationship between amyloid aggregation and both oxidative stress36 and acetylcholinesterase activity37 during AD progression.
Oxidative stress is related to the neuropathological manifestations of AD and implies an increased level of reactive oxygen species (ROS)38,39 and the abnormal homeostasis of bioactive metals40. Otherwise, acetylcholinesterase catalyzes acetylcholine conversion, which is related to the cholinergic cascade and cholinergic neuron loss in AD pathology41.
In an effort to target those processes, antioxidant compounds (e.g. resveratrol) have shown a role in AD prevention42 or as a supportive treatment43. Furthermore, many AD drugs inhibit acetylcholinesterase activity, aiming to increase acetylcholine levels in the brain44. However, these drugs have a limited effect and generate collateral effects45.
Therefore, with a positive prediction from the in silico investigation, we screened the effect of the kefir fractions in vitro. Both antioxidant and anti-acetylcholinesterase properties were confirmed using the FRAP and acetylcholinesterase inhibition assays. To the best of our knowledge, this is the first work to report the anti-acetylcholinesterase activity of a kefir-produced molecule.
In order to verify the in vivo effects of peptides from kefir, we used the D. melanogaster AD-like model. For that, we first provided a confirmation of the model, in which human BACE and APP are overexpressed in fly neurons by the pan-neural driver elav-Gal4.
In the functional assays, the flies displayed a motor deficit starting at 10–13 d.p.e, which was correlated with a increase in amyloid relative level, demonstrated by amyloidogenic quantification, and severe neurodegeneration based on the histopathological analysis. This has been previously described for a similar model13,46,47 and our previous data with the model used here48, which indicates the suitability of the model.
With a validated model in hand, we treated AD-like flies with kefir fractions. Flies treated with the WSF and < 10 kDa fractions (both at 0.25 mg/mL) displayed better climbing ability in relation to untreated flies. However, the < 10 kDa fraction had an apparently stronger influence on this process, probably due to its higher peptide concentration due to the serial filtration process. However, although the > 10 kDa fraction improved the FRAP results, the neurodegeneration index and relative Aβ peptides, it was not able to inhibit acetylcholinesterase. Dose dependency was not observed here, which is possible since both fractions were constituted of a mix of peptides and polypeptides. The higher tested concentration (0.5 mg/mL) likely contained bioactive peptides against other targets can act in another sites. Again, only the lower concentration of the > 10 kDa fraction was able to generate a significant decrease in AChE activity.
To investigate this effect, first the peptides present in the < 10 kDa fraction were identified by LC–MS/MS. Then, the 3D peptides were predicted and finally molecular dockings was performed to identify potentially bioactive peptides with AChE, BACE1, and Aβ peptides as the targets. AChE has two binding sites: one is a PAS located at the entrance of the active site gorge, and the other is a CAS located at the base of the active site gorge49; ligands of PAS or CAS can inactivate this enzyme50. The binding of the VAPFPEVFG peptide at the PAS could be enough to promote the inhibition of acetylcholinesterase. Our docking data are only a tentative explanation for the characteristics and effects of kefir peptides. The real effects observed in vivo in the AD-like model were likely promoted by a complex mix of peptides.
The BACE1 molecule is formed of three portions: an extracellular N-terminal domain, a transmembrane domain and a C-terminal cytosolic domain. Its cleavage site is between the N- and C-terminal regions, characterized by an aspartate catalytic dyad (Asp32 and Asp228, highlighted in yellow in Fig. 5a,b)51. Near this region, there is a flexible flap (highlighted in green in Fig. 5a,b), perpendicular to the active site. This flexible flap can exist in either an open or closed conformation, and in that way help or hinder the access of a molecule to the enzyme active site52,53. Between the flap and the active site, there is also a space in the BACE1 structure that can be reached by a molecule so that it can access the catalytic dyad54.
By analyzing the 3D structure of BACE1 and the peptide VPPFLQPEV (Table 1 docking of smaller global energy value, Fig. 5a,b), it was possible to verify that it did not interact with the BACE1 active site. Despite this, the peptide interacted with ALCA, and was able to alter its conformation, strategically located between the large N- and C-terminal portions of BACE1. This peptide position could block the access of other molecules (substrates) to the enzyme active site, thereby inhibiting its action. As a consequence, BACE1 could lose its capacity to cleave APP, consequently reducing Aβ peptide production and amyloid plaque accumulation. In this way, it could be possible to decrease the progression of neurodegeneration through the amyloidogenic pathway.
The amyloidogenic pathway results in the production of Aβ peptides of distinct sizes, depending on the γ-secretase cleavage region. Amongst those, the 42 amino acid peptide (Aβ42) is the most neurotoxic55,56. Based on that, we also evaluated molecular docking between the bioactive peptide VYPFPGPIPN and amyloid plaques generated by the Aβ42 peptide.
This peptide has a unique conformation, composed of three β-sheets that cover the residues 12–18 (β1), 24–33 (β2) and 36–40 (β3)57. This tertiary fold is responsible for the toxicity and aggregation of this peptide, which results in β-amyloid plaque formation in AD. Based on the molecular docking analysis between the peptide VYPFPGPIPN and amyloid plaques (Fig. 5c,e), we observed an interaction between those molecules, and a consequent conformational change in the amyloid plaque, which was highlighted by the opening of its tertiary fold (Fig. 5e). This alteration in β-amyloid plaque conformation could reduce its toxicity, thus neutralizing the negative effects of its aggregation and therefore attenuating the effects of AD.
The present study extends the existing literature by providing evidence that peptides derived from cow milk kefir can modulate the AD phenotype in AD-like flies by decreasing the relative β-amyloid level in the brain. Consequently, this intervention decreases neuronal tissue damage, improves motor ability and decreases acetylcholinesterase activity. In summary, our study was able to identify bioactive peptides present in kefir (< 10 kDa fraction) and to predict their potential use as an alternative adjuvant treatment for AD or as a drug prototype.
Methods
Samples
Kefir grains were obtained as a donation from the local population in Uberlândia, Brazil, inoculated (4% w/v) in UHT (ultra-high temperature) whole cow milk for fermentation process and left for 24 h at room temperature in a glass container. A genetic fingerprint was previously published by our group13. The fermented product was collected by removing the grains by filtration, followed by centrifugation at 4 °C, 4900×g for 10 min. The resulting supernatant went through a series of filtering processes, where we obtained the three fractions that were further tested.
The pellet was discarded, and the supernatant was re-centrifuged, generating the kefir water-soluble fraction (WSF). This was then filtered using a vacuum pump and passed through a 0.22 µm membrane. Finally, this filtrate was passed through a 10 kDa Amicon column, separating peptides by size: larger than 10 kDa (referred to as > 10 kDa) and smaller than 10 kDa (< 10 kDa).
Proteopeptidomics
In preparation for shotgun proteomics, the samples were reduced with 100 mM dithiothreitol (DTT) and alkylated with 0.5 M iodoacetamide. Samples were then digested with trypsin (0.01 μg/μL), except for the < 10 kDa peptide fraction, which was obtained from Amicon filtering. A desalination step was performed with a ZipTips C18 column (Millipore, Billerica, MA, United States).
Shotgun proteomics and peptidomics were performed by liquid chromatography–electrospray ionization–quadrupole–time of flight-mass Spectrometry (LC–ESI–Q–TOF–MS) using the 6520B instrument from Agilent. An AdvanceBio Peptide Mapping column (Agilent) was used (2.1 mm internal diameter, 10 cm long, 2.7 µm particles). In the mobile phase, water (A) and acetonitrile (B) were acidified with formic acid (0.1% v/v), using the gradient: 2% B (0 min), 2% B (10 min), 15% B (40 min), 50% B (150 min), 70% B (200 min), 98% B (220 min), 98% B (300 min), 100% B (301 min) and 100% B (400 min) at 400 µL/min. For ionization, a nebulizer pressure of 45 psi was used, with drying gas at 8 L/min (325 °C). 4 kV energy was applied to the capillary.
Spectrum Mill software (Agilent) was used for data analysis, using the following Uniprot databases: (1) “milk AND bovine” (69,760 results in February 2021); (2) “Lactobacillus” (1,250,616 results in February 2021) and (3) “Acetobacter” (185,991 results in February 2021). Carbamidomethylation was set as fixed with and. Maximum missed cleavages were selected in two for trypsin. The precursor mass error and the fragments at 20 ppm, product mass tolerance at 50 ppm, and maximum ambiguous precursor charge at 3.
For de novo analysis, the Sherenga module of the Spectrum Mill software (Agilent) was used.
In silico physicochemical parameters, toxicity, and bioactivity prediction of obtained peptides
ToxinPred (https://webs.iiitd.edu.in/raghava/toxinpred/algo.php) was used to calculate the physicochemical characteristics of each characterized peptide and the expected toxicity level, while the Peptide Property Calculator from Innovagen (http://www.innovagen.com/proteomics-tools) was used to analyze solubility. For the bioactivity prediction, PeptideRanker (http://distilldeep.ucd.ie/PeptideRanker/) was used. Finally, the Milk Bioactive Peptide Database (http://mbpdb.nws.oregonstate.edu/) was used to determine the origin of peptides from milk protein and their putative biological functions. The peptides with a rank above 0.5 were considered as having potential bioactivity.
In vitro fraction evaluations
Total antioxidant activity
Total antioxidant capacity was evaluated by FRAP (ferric reducing antioxidant power) assay20. This method consists of evaluating the capacity if a compound reduction Fe3+ to Fe2+ Kefir fractions (WSF and both Amicon samples) were solubilized in distilled water at 500 µg/mL. Ascorbic acid was used at the same concentration as the positive control and sodium acetate buffer was used as the blank.
For the assay, 250 µL of FRAP reagent (10× sodium acetate 0.3 M, TPTZ (2,3,5-triphenyltetrazolium chloride) 10 mM, 1 volume of ferric chloride 20 mM) were mixed with 10 µL of each sample and 25 µL of MilliQ Water. The reaction progressed for 6 min at 37 °C. The respective absorbances were measured at 593 nm on a spectrophotometer. The antioxidant capacity of each sample was determined by the construction of an analytical curve built with Trolox (6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid).
Acetylcholinesterase inhibition assay
The acetylcholinesterase inhibition assay was based on the one described by Rhee58. In this assay, three Tris–HCl buffers were used: (I) standard Tris–HCl buffer (pH 8); (II) Tris–HCl with bovine serum albumin 1% w/v; and III) Tris–HCl with NaCl 0.6% w/v and MgCl2 0.4% w/v. The enzyme was diluted (0.2 U/mL) in buffer I.
The solutions were prepared in the following way: the enzyme was diluted to 0.2 U/mL in buffer I, DTNB 0.1% was diluted in buffer III, acetylcholine iodide solution (substrate) 0.4% v/v was diluted in Milli-Q water, and the inhibitor Galantamine (positive control) diluted in Milli-Q water.
For the assay, 25 µL of each sample were added to 125 µL of DTNB solution, 50 µL of buffer II, 25 µL of acetylcholine iodide solution and 25 µL of acetylcholinesterase solution. Using a spectrophotometer, the absorbance was measured at 405 nm for 20 min at 30 °C. Acetylcholinesterase inhibition was calculated using the following equation:
In vivo experiments
Drosophila melanogaster genetics
Fly stocks were obtained from the Bloomington Stock Center: w1118 (stock# 3605), UAS-BACE1, UAS-APP (#33797) and elav-GAL4 (#5146). Flies were kept in an incubator at 25 °C in a 12 h:12 h light:dark cycle. Unless otherwise stated, flies were reared on cornmeal medium (soy powder 0.01%, glucose 7.2%, agar 0.6%, cornmeal 0. 073%, yeast 0.018%, nipagin 0.06% and acid solution 0.05% w/v).
AD-like flies expressing human BACE and APP pan-neuronally were obtained by crossing elav-GAL4 and UAS-BACE, UAS-APP flies. Resulting pupae were selected according to phenotype—without any genetic marker, as tubby—ensuring that the resulting fly genotype is elav-Gal4/+; UAS-BACE, UAS-APP/+ (AD-like model).
Treatments
Fly treatment was administered via the food and began at 0–3 days post eclosion. All kefir samples were solubilized at 0.5 and 0.25 mg/mL. For this, 5 mL of sample was mixed into 1 g of enriched mashed potato medium (75% instant mashed potato, 15% yeast extract, 9.3% glucose, and 0.07% nipagin). The treatment medium was changed every other day to ensure fresh exposure to the samples. Control flies were fed with enriched potato powder medium solubilized with distilled water only. For all experiments, only male flies were used.
Rapid iterative negative geotaxis assay (RING)
The rapid iterative negative geotaxis assay (RING) was adapted from Gargano et al.59 and used to evaluate the effect of different types of treatment on fly locomotor ability. To ensure an AD-like phenotype, these flies were compared to control flies (elav-Gal4/+). Groups of 30 AD-like flies (in triplicate) for each treatment (WSF, 0.22 µm, and both Amicon conditions) were transferred to clean vials (9.5 cm × 2.5 cm) and placed in a custom vial holder. Flies had their behavior assessed 5 and 10 days after treatment. Before testing, flies were exposed to light and kept in a silent environment for 20 min, to acclimate. For the assay, the holder was hit three times on the bench, and the flies were given 4 s to climb 5 cm. This was repeated five times. The procedure was recorded, and the video was analyzed using QuickTime Player 7.7.9 software. The average climbing percentage was calculated as the percentage of flies of each group that reached the 5 cm mark 120 frames (4 s) after the holder touched the bench.
Total antioxidant activity in vivo
To evaluate the total antioxidant activity for each treatment, the modified FRAP method was used. Ten fly heads (in triplicate) after 10 days of treatment were homogenized with PBS and centrifuged (2 min, 1000×g at 4 °C). For the assay, 10 µL of each supernatant was mixed with 50 µL of FRAP reagent 1:1 in distilled water. Antioxidant activity was evaluated as previously described for the in vitro analysis.
Acetylcholinesterase activity assay
For the assay, homogenized fly head solutions were made as described above. Then, 25 µL of the solution was added to 50 µL of Tris–HCl buffer (bovine serum albumin 1% w/v), 125 µL of DNTB, 25 µL of acetylcholine iodine solution and 25 µL of acetylcholinesterase (0.2 U/mL) (based on Ellman’s protocol)60. Sample absorbance was measured at 405 nm at 30 °C for 20 min at 30 s intervals.
Amyloid quantification
The amyloid content was assessed using Thioflavin T (ThT), a benzothiazole dye that exhibits enhanced fluorescence upon binding to amyloid fibrils. Fly heads from elav-Gal4/+ (control) and AD-like flies (a pool of 10 heads each, in triplicate) were collected, frozen in liquid nitrogen, and kept at − 80 °C until the next step. Fly heads were then kept on ice and homogenized in PBS 1×, then centrifuged (2 min, 10,000×g at 4 °C). The supernatants were collected and used for amyloid quantification and Bradford protein levels in technical triplicates. In a protocol modified from Westfall et al.61 supernatants were incubated with the ThT working solution (20 µM) for 20 min under agitation. Fluorescence was measured at 450 nm excitation/482 nm emission and normalized to ThT only samples. Total fluorescence was corrected to the total protein content of each sample and to elav-Gal4/+ (control flies) fluorescence levels. This additional step was needed as in vivo samples could present autofluorescence.
Histopathological analysis
Ten flies were used from each treatment group, using the optimal concentration obtained in previous experiments. All flies were analyzed at 10–13 days post eclosion (d.p.e.) and elav-Gal4/+ was used as the control. Fly heads were transferred to 4% formaldehyde in sodium phosphate buffer 0.1 M pH 7.2 for 16 h at 4 °C. The samples were then dehydrated in a graded ethanol series (70, 80, 90, and 95%) and transferred to methanol for 16 h at 4 °C. Next, they were embedded in HistoResin (Leica) and 3 µm thick slices were stained with hematoxylin and eosin, analyzed and photographed with a light photomicroscope. Neuropile images were used to calculate the neurodegenerative index as normal to low, moderate or severe, according to vacuolar lesions13,22.
Molecular docking
Putative bioactive components were used for docking analysis. The 3D peptide structures were created using PEP-FOLD 2.0. The Protein Data Bank (PDB) files for BACE (3TPJ), acetylcholinesterase (AChE-3LII) and 42-Residue Beta-Amyloid Fibril (2MXU), were retrieved from PDB. The docking between the peptides and these enzymes was performed using PathDocking. The best model was chosen based on both global energy and atomic contact energy contribution to the global binding energy.
Statistical analysis
Data analysis was performed using GraphPad Prism 8. A priori, we evaluated the normal distribution of the data by the D’Agostino and Pearson test. Groups were compared through a t-test with an established significance level of p < 0.05.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request. The raw data from mass spectrometry analyses were deposited in PRIDE access number: PXD034190 (fraction < 10 kDa digested with trypsin), PXD034189 (fraction > 10 kDa digested with trypsin) and PXD034148 (intact fraction < 10 kDa).
References
Talwar, P. et al. Dissecting complex and multifactorial nature of Alzheimer’s disease pathogenesis: A clinical, genomic, and systems biology perspective. Mol. Neurobiol. 53, 4833–4864 (2016).
Weller, J., Budson, A., Portelius, E. & Reddy, H. Current understanding of Alzheimer’s disease diagnosis and treatment. F1000Research 7, 1161 (2018).
Nichols, E. et al. Estimation of the global prevalence of dementia in 2019 and forecasted prevalence in 2050: An analysis for the Global Burden of Disease Study 2019. Lancet Public Health 7, e105–e125 (2022).
Breijyeh, Z. & Karaman, R. Comprehensive review on Alzheimer’s disease: Causes and treatment. Molecules (Basel) 25, 5789 (2020).
Jellinger, K. A. Neuropathological assessment of the Alzheimer spectrum. J. Neural Transm. 127, 1229–1256 (2020).
Lane, C. A., Hardy, J. & Schott, J. M. Alzheimer’s disease. Eur. J. Neurol. 25, 59–70 (2018).
Sharma, V. K. et al. Dysbiosis and Alzheimer’s disease: A role for chronic stress? Biomolecules 11, 678 (2021).
Kowalski, K. & Mulak, A. Brain-gut-microbiota axis in Alzheimer’s disease. J. Neurogastroenterol. Motil. 25, 48–60 (2019).
Bonfili, L. et al. Microbiota modulation counteracts Alzheimer’s disease progression influencing neuronal proteolysis and gut hormones plasma levels. Sci. Rep. 7, 1 (2017).
Wong, C. B., Kobayashi, Y. & Xiao, J. Probiotics for preventing cognitive impairment in Alzheimer’s disease. Gut Microbiota Brain Axis. https://doi.org/10.5772/INTECHOPEN.79088 (2018).
Nielsen, B., Gürakan, G. C. & Ünlü, G. Kefir: A multifaceted fermented dairy product. Probiot. Antimicrob. Proteins 6, 123–135 (2014).
Plessas, S. et al. Microbiological exploration of different types of kefir grains. Ferment 3, 1 (2016).
Batista, L. L. et al. Kefir metabolites in a fly model for Alzheimer’s disease. Sci. Rep. 11, 11262 (2021).
Tsuda, L. & Lim, Y.-M. Alzheimer’s disease model system using drosophila. Adv. Exp. Med. Biol. 1076, 25–40 (2018).
Chakraborty, R. et al. Characterization of a drosophila Alzheimer’s disease model: Pharmacological rescue of cognitive defects. PLoS ONE 6, e20799 (2011).
Jeon, Y., Lee, J. H., Choi, B., Won, S. Y. & Cho, K. S. Genetic dissection of Alzheimer’s disease using Drosophila models. Int. J. Mol. Sci. 21, 884 (2020).
Jeibmann, A. & Paulus, W. Drosophila melanogaster as a model organism of brain diseases. Int. J. Mol. Sci. 10, 407–440 (2009).
Prüßing, K., Voigt, A. & Schulz, J. B. Drosophila melanogaster as a model organism for Alzheimer’s disease. Mol. Neurodegener. 8, 35 (2013).
Tue, N. T., Dat, T. Q., Ly, L. L., Anh, V. D. & Yoshida, H. Insights from Drosophila melanogaster model of Alzheimer’s disease. Front. Biosci. Landmark 25, 134–146 (2020).
Benzie, I. F. F. & Strain, J. J. The ferric reducing ability of plasma (FRAP) as a measure of “antioxidant power”: The FRAP assay. Anal. Biochem. 239, 70–76 (1996).
Khurana, R. et al. Mechanism of thioflavin T binding to amyloid fibrils. J. Struct. Biol. 151, 229–238 (2005).
Cao, Y., Chtarbanova, S., Petersen, A. J. & Ganetzky, B. Dnr1 mutations cause neurodegeneration in Drosophila by activating the innate immune response in the brain. Proc. Natl. Acad. Sci. 110, E1752–E1760 (2013).
Tung, M.-C. et al. Kefir peptides alleviate high-fat diet-induced atherosclerosis by attenuating macrophage accumulation and oxidative stress in ApoE knockout mice. Sci. Rep. 10, 1–15 (2020).
Pimenta, F. S. et al. Mechanisms of action of kefir in chronic cardiovascular and metabolic diseases. Cell. Physiol. Biochem. 48, 1901–1914 (2018).
Hamida, R. S. et al. Kefir: A protective dietary supplementation against viral infection. Biomed. Pharmacother. 133, 110974 (2021).
Bourrie, B. C. T., Richard, C. & Willing, B. P. Kefir in the prevention and treatment of obesity and metabolic disorders. Curr. Nutr. Rep. 9, 184–192 (2020).
Azizi, N. F. et al. Kefir and its biological activities. Foods 10, 1210 (2021).
Barao, C. E. et al. Growth kinetics of kefir biomass: Influence of the incubation temperature in milk. Chem. Eng. Trans. 75, 499–504 (2019).
Londero, A., Hamet, M. F., De Antoni, G. L., Garrote, G. L. & Abraham, A. G. Kefir grains as a starter for whey fermentation at different temperatures: Chemical and microbiological characterisation. J. Dairy Res. 79, 262–271 (2012).
Dallas, D. C. et al. Peptidomic analysis reveals proteolytic activity of kefir microorganisms on bovine milk proteins. Food Chem. 197, 273–284 (2016).
Amorim, F. G. et al. Identification of new bioactive peptides from Kefir milk through proteopeptidomics: Bioprospection of antihypertensive molecules. Food Chem. 282, 109–119 (2019).
Fan, M. et al. Isolation and identification of novel casein-derived bioactive peptides and potential functions in fermented casein with Lactobacillus helveticus. Food Sci. Hum. Wellness 8, 156–176 (2019).
Marcone, S., Belton, O. & Fitzgerald, D. J. Milk-derived bioactive peptides and their health promoting effects: A potential role in atherosclerosis. Br. J. Clin. Pharmacol. 83, 152–162 (2017).
Nagpal, R. et al. Bioactive peptides derived from milk proteins and their health beneficial potentials: An update. Food Funct. 2, 18–27 (2011).
Kaur, D. et al. Multifaceted Alzheimer’s disease: Building a roadmap for advancement of novel therapies. Neurochem. Res. https://doi.org/10.1007/S11064-021-03415-W (2021).
Zhao, Y. & Zhao, B. Oxidative stress and the pathogenesis of Alzheimer’s disease. Oxid. Med. Cell. Longev. https://doi.org/10.1155/2013/316523 (2013).
Nalivaeva, N. & Turner, A. AChE and the amyloid precursor protein (APP)—Cross-talk in Alzheimer’s disease. Chem. Biol. Interact. 259, 301–306 (2016).
Tönnies, E. & Trushina, E. Oxidative stress, synaptic dysfunction, and Alzheimer’s disease. J. Alzheimers Dis. 57, 1105–1121 (2017).
Huang, W., Zhang, X. & Chen, W. Role of oxidative stress in Alzheimer’s disease. Biomed. Reports 4, 519–522 (2016).
Birla, H., Minocha, T., Kumar, G., Misra, A. & Singh, S. K. Role of oxidative stress and metal toxicity in the progression of Alzheimer’s disease. Curr. Neuropharmacol. 18, 552–562 (2020).
Ferreira-Vieira, T. H., Guimaraes, I. M., Silva, F. R. & Ribeiro, F. M. Alzheimer’s disease: Targeting the cholinergic system. Curr. Neuropharmacol. 14, 101 (2016).
Grodzicki, W. & Dziendzikowska, K. The role of selected bioactive compounds in the prevention of Alzheimer’s disease. Antioxidants 9, 229 (2020).
Sousa, J. C. E., Santana, A. C. F. & Magalhães, G. J. P. Resveratrol in Alzheimer’s disease: A review of pathophysiology and therapeutic potential. Arq. Neuropsiquiatr. 78, 501–511 (2020).
Forlenza, O. V. Tratamento farmacológico da doença de Alzheimer. Arch. Clin. Psychiatry (São Paulo) 32, 137–148 (2005).
Yiannopoulou, K. G. & Papageorgiou, S. G. Current and future treatments for Alzheimer’s disease. Ther. Adv. Neurol. Disord. 6, 19 (2013).
Wang, X. et al. Effects of curcuminoids identified in rhizomes of Curcuma longa on BACE-1 inhibitory and behavioral activity and lifespan of Alzheimer’s disease Drosophila models. BMC Complement. Altern. Med. 14, 88 (2014).
Chiu, W. Y. V. et al. GULP1/CED-6 ameliorates amyloid-β toxicity in a Drosophila model of Alzheimer’s disease. Oncotarget 8, 99274–99283 (2017).
da Costa Silva, J. R. et al. Differential gene expression by RNA-seq during Alzheimer’s disease-like progression in the Drosophila melanogaster model. Neurosci. Res. https://doi.org/10.1016/J.NEURES.2022.02.003 (2022).
Yu, Z. et al. Identification and molecular docking study of fish roe-derived peptides as potent BACE 1, AChE, and BChE inhibitors. Food Funct. 11, 6643–6651 (2020).
Dvir, H., Silman, I., Harel, M., Rosenberry, T. L. & Sussman, J. L. Acetylcholinesterase: From 3D structure to function. Chem. Biol. Interact. 187, 10–22 (2010).
Hong, L. et al. Structure of the protease domain of memapsin 2 (β-secretase) complexed with inhibitor. Science 290, 150–153 (2000).
Hong, L. & Tang, J. Flap position of free memapsin 2 (β-secretase), a model for flap opening in aspartic protease catalysis†,‡. Biochemistry 43, 4689–4695 (2004).
James, M. N., Sielecki, A., Salituro, F., Rich, D. H. & Hofmann, T. Conformational flexibility in the active sites of aspartyl proteinases revealed by a pepstatin fragment binding to penicillopepsin. Proc. Natl. Acad. Sci. 79, 6137–6141 (1982).
Shimizu, H. et al. Crystal structure of an active form of BACE1, an enzyme responsible for amyloid β protein production. Mol. Cell. Biol. 28, 3663–3671 (2008).
Ray, B. et al. Rivastigmine modifies the α-secretase pathway and potentially early Alzheimer’s disease. Transl. Psychiatry 10, 1–17 (2020).
Luheshi, L. M. et al. Systematic in vivo analysis of the intrinsic determinants of amyloid β pathogenicity. PLoS Biol. 5, e290 (2007).
Xiao, Y. et al. Aβ(1–42) fibril structure illuminates self-recognition and replication of amyloid in Alzheimer’s disease. Nat. Struct. Mol. Biol. 22, 499–505 (2015).
Rhee, I. K., Van De Meent, M., Ingkaninan, K. & Verpoorte, R. Screening for acetylcholinesterase inhibitors from Amaryllidaceae using silica gel thin-layer chromatography in combination with bioactivity staining. J. Chromatogr. A 915, 217–223 (2001).
Gargano, J. W., Martin, I., Bhandari, P. & Grotewiel, M. S. Rapid iterative negative geotaxis (RING): A new method for assessing age-related locomotor decline in Drosophila. Exp. Gerontol. 40, 386–395 (2005).
Ellman, G. L., Courtney, K. D., Andres, V. & Featherstone, R. M. A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem. Pharmacol. 7, 88–95 (1961).
Westfall, S., Lomis, N. & Prakash, S. A novel synbiotic delays Alzheimer’s disease onset via combinatorial gut-brain-axis signaling in Drosophila melanogaster. PLoS ONE 14, e0214985 (2019).
Acknowledgements
The authors are thankful to the Institute of Cell Biology for providing access to the Light Microscope and Software LAS EZ and to the Pathology Laboratory of the Faculty of Odontology of the Federal University of Uberlândia for the microscope sections. We are grateful for the technical and material support of Professor Dr. Luiz Ricardo Goulart, from the Biotechnology Institute of the University of Uberlândia, in memoriam.
Funding
This research was funded by the Conselho Nacional Científico e Tecnológico do Brasil (CNPq, 312812/2021-3 and 303667/2021-4), the Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG, APQ-02766-17 and PPM-00503-18) and the Coordenadoria de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). Federal University of Uberlândia provided physical conditions for experimental procedures.
Author information
Authors and Affiliations
Contributions
S.M.M. Conception, design, development of the methodology, analysis and interpretation of data, writing and revision of the manuscript. L.L.B.; H.C.G.S.; R.R.F.; M.H.S.; T.S.R.; L.I.V.C.; M.M.M.; G.V.; Performed investigation. F.S.E. technical and material support. M.V.S.e.C.U.V. supervised the project. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Malta, S.M., Batista, L.L., Silva, H.C.G. et al. Identification of bioactive peptides from a Brazilian kefir sample, and their anti-Alzheimer potential in Drosophila melanogaster. Sci Rep 12, 11065 (2022). https://doi.org/10.1038/s41598-022-15297-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-15297-1
This article is cited by
-
Development of nutri-functional paneer whey-based kefir drink
Journal of Food Science and Technology (2025)
-
Unveiling the Brazilian kefir microbiome: discovery of a novel Lactobacillus kefiranofaciens (LkefirU) genome and in silico prospection of bioactive peptides with potential anti-Alzheimer properties
BMC Genomics (2024)
-
Green synthesis of silver nanoparticle using pollen extract from Tetragonisca angustula a stingless bee
Discover Nano (2024)
-
De novo antioxidant peptide design via machine learning and DFT studies
Scientific Reports (2024)
-
Nutrients and polyphenols-rich Sorghum bicolor genotypes as complementary therapy for Alzheimer’s disease
Phytochemistry Reviews (2024)