Abstract
Patients with oral squamous cell carcinoma (OSCC) present significant alterations in their saliva proteome. We have used the shotgun Phage Display (PD) technology to identify candidate proteins that were upregulated in saliva of OSCC by selecting ligands to salivary proteins from a single-chain variable fragment (scFv) PD combinatorial library. After two selection cycles, the highly reactive clone scFv-D09 was able to distinguish saliva of OSCC patients from healthy subjects by enzyme-linked immunosorbent assay (ELISA) with sensitivity and specificity of 96.67%. Additionally, the scFv-D09 clone presented a positive immunostaining for invasive malignant epithelial cells in the connective tissue, keratin pearls in the OSCC, and ducts of salivary glands. We have further identified the target protein as the tropomyosin alpha-4 chain (TPM4) by two-dimensional polyacrylamide gel electrophoresis and mass spectrometry, and its binding to the scFV-D09 was demonstrated by bioinformatics. Briefly, we have identified TPM4 as upregulated salivary protein in patients with OSCC, which plays a central role in stabilizing cytoskeleton actin filaments, probably linked with tumor tissue remodeling. Long-term longitudinal studies are needed to validate TPM4 as a potential marker of a malignant process.
Similar content being viewed by others
Introduction
Oral cancer (OC) encompasses a heterogeneous group of cancers affecting oral tissues. Statistics from GLOBOCAN-IARC (2018) point out that oral and lip cancers are the 18th type of human cancer in frequency, reaching 2.0% of the new cases among 36 human cancers combined, presenting a variable mortality rate lower than 65%1,2. In Brazil, oral cancers is the fifth type of cancer in men with an incidence of 5.2% of all nonmelanoma human cancers, with a mortality rate of approximately 44%3. The oral squamous cell carcinoma (OSCC) represents more than 95% of oral cancer tissues, and its incidence and mortality rates are representative of the oral cancer group1,2. The main risk factors for OSCC is usually diagnosed in advanced stages of malignancy by visual inspection of lesions followed by biopsy4. Moreover, the biopsy still remains the gold standard in the final diagnosis of the OSCC5.
Studies have shown that OSCC patients present significant modifications in the saliva proteome6,7. Saliva is a clinically informative biological fluid; its direct contact with oral lesions makes it highly useful for diagnosis and monitoring oral cancers and systemic diseases7,8. Furthermore, saliva is a convenient and easy sample to obtain, which can be collected with a noninvasive and cost-effective method9.
The search for salivary biomarkers for OSCC have included DNA, RNA and proteins10,11,12,13. Several salivary protein markers have been detected at high concentrations in saliva of OSCC, for example, interleukin 610 and 811, epidermal growth factor receptor (EGFR)12, Cyfra 21.114, catalase15, tumor necrosis factor (TNF-α)10, the peptide defensin- 116 and S100A8 acid protein17, which may be considered putative biomarkers for OSCC diagnosis, but none of them were validated.
Combinatorial technologies, such as Phage Display, have also led to the detection of specific proteins differentially expressed between cancer and non-cancer samples18,19,20, and this technology is highly desirable since it allows selection of interesting combinatorial antibodies targeting specific antigens from different samples21.
Here, we report an exploratory strategy that identifies a highly expressed protein in saliva of OSCC patients using an scFv orignated from a combinatorial phage library selection. This protein target was Tropomyosin alpha-4 chain, which was characterized by immunoassays and mass spectrometry, and these findings is discussed herein.
Results
Selection, DNA sequencing and bioinformatics analysis
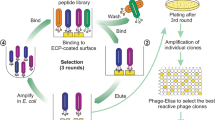
A scFv PD library derived from head and neck cancer19 was used to select total salivary protein ligands from the saliva of OSCC patients by prior subtraction from healthy individuals. After the second round of panning, clones were grown, and the soluble expression of the scFv fragments were verified by dot blot, and the clone with the highest expression, scFv-D09, was amplified and and its DNA was characterized by sequencing. The scFv was then purified by HPLC for additional characterization. The predicted amino-acid sequence for both heavy and light chains of the scFv-D09 clone is shown in Fig. 1A, including all three complementarity determining regions (CDRs) and conserved framework regions (FWRs). The clone was submitted to in silico prediction tools (PyMOL and RaptorX) for its 3D structural analyses (Fig. 1B,C).
Single-chain fragment variable antibody (scFv). (A) The amino acid sequence of scFv-D09 clone with appropriate regions for framework and complementarity-determining regions (CDRs) residues, variable light chain (VL) and the variable heavy chain (VH) domain. (B,C) The 3D structure of scFv molecule and predicted antigen- binding site (CDRs), both analyzed by the RaptorX and PyMOL online tool.
scFv-D09 detects an antigen present in saliva of patients with OSCC
Total protein of saliva from OSCC and healthy subjects’ group was immobilized in high affinity microtiter plates for scFv-D09 detection. Data of reactivity index demonstrated a significant discrimination between OSCC patients in relation to healthy subjects (P < 0.0001) (Fig. 2A). In OSSC group (29/30) tested positive for the scFv and one healthy subject was diagnosed as positive. Based on the ROC curve analyses, the cut-off value chosen for scFv-D09 was 0.0625 (Fig. 2B), and both sensitivity and specificity were 96.67% (82.78–99.92). The test presented a very high efficiency with a positive likelihood ratio of 29.0, and area under the curve of 0.9794. There is no correlation between the clinicopathologic characteristics and the Elisa absorbance of oral cancer patients.
ELISA using scFv to detect salivary proteins. (A) Reactivity indices of saliva from individuals with Oral squamous cell carcinoma (OSCC, n = 30) and healthy subjects (Controls, n = 30) analyzed by enzyme-linked immunosorbent assay (ELISA). Scatter dot-plots with ELISA reactivity index (RI) according to selected cut-off, mean with standard deviation; Mann Whitney test (*P < 0.0001). (B) ROC curve showing sensitivity (Se), specificity (Sp), positive likelihood ratio (LR+) and area under the curve (AUC) (P < 0.001).
scFv-D09 recognizes OSCC tissue
Immunohistochemical analysis of scFv-D09 antibody was performed in OSCC (n = 10) and control tissues (n = 10). It revealed a positive staining in keratin pearls, invasion of malignant epithelial cells in the connective tissue in OSCC and ducts of the salivary glands, as demonstrated in Fig. 3 panels A, B and C, respectively. No labeling was detected in negative control, tissue without scFv-D09- secondary Ab alone (Fig. 3D–F) and control tissue, mucocele, a benign cystic lesion with scFv-D09 (Fig. 3G–I).
Immunohistochemistry with scFv-D09 antibody in oral tissue. Immunostaining of the scFv-D09 antibody demonstrates reaction in: (A) keratin pearls of OSCC, (B) invasion of malignant epithelial cells in the connective tissue in OSCC and (C) ducts of the salivary glands. Negative staining for control without scFv- secondary Ab alone was observed in (D) keratin pearls, (E) invasion of malignant epithelial cells in the connective tissue and (F) ducts of the salivary glands. No labeling with scFv D09 was detected in control tissue- mucocele- a benign cystic lesion (G,H,I).
Identification of Tropomyosin alpha-4 chain protein as target
The two-dimensional polyacrylamide gel electrophoresis (2DE) method was performed to separate pools of protein extracts from OSCC tissues. Figure 4 shows the representative proteomic profile for oral carcinoma in panel A, and panel B represents the Western blot detected spot after incubation with scFv-D09 antibody. As shown (arrow) the scFv D09 antibody recognized only one spot, which was further excised from 2D electrophoresis gel and identified by mass spectrometry analysis as tropomyosin alpha-4 chain protein (Fig. 4B). The theoretical molecular mass/pI values for TPM4 was 28,522 kDa/4.67, with an excellent fit with its corresponding spot in the 2-DE gel (Fig. 4A), presenting a sequence coverage of 14% (UniProtKB accession number P67936). Docking between scFv and TPM4 protein was also performed to determine possible binding regions, and the most stable 3D structure of the TPM4-scFv complex was futher analyzed by PyMOL, which identified the CDRs that were possibly responsible for the scFv binding to the TPM4 epitope (Fig. 4C). The original gel, membrane and western blot images are shown in the Supplementary Information.
Identification of Tropomyosin alpha 4 chain in OSSC. (A) Two-dimensional electrophoresis gel of oral squamous cell carcinoma. The sizes of the molecular weight markers (MW) are shown on the left side of panel and the pH range was 4–7. (B) Western blot analysis of the scFv-D09 antibody with Tropomyosin alpha 4 chain detection (arrow). (C) scFv-D09 and Tropomyosin alpha 4 chain binding was performed using Patchdock and PyMOL online tools. The scFv predominantely binds to the N-portion of the protein.
Discussion
This study describes the identification of Tropomyosin alpha-4 (TPM4) chain as an antigen in saliva of OSCC patients. Traditionally, the search for biomarkers in human saliva have been done through detection of differentially expressed proteins in 2D gels followed by mass spectrometry analysis15,22,23. Here, we have used a recombinant PD library with a subtractive proteomic selection strategy as a discovery tool to identify proteins that were upregulated in saliva of OSCC patients. The subtractive approach against salivary proteins of healthy subjects was followed by a positive selection against total salivary proteins of OSCC patients in order to determine functional epitopes in the active disease, minimizing cross-reactions, and favoring the detection of highly specific antigens.
ELISA results indicated that the scFv-D09 was able to discriminate the saliva of cancer patients from healthy subjects. In addition, it revealed a positive staining for invasion of malignant epithelial cells in the connective tissue and keratin pearls in OSCC. Mass spectrometry analysis of the immunostained spot with scFv-D09 in the 2D gel western blot led to the identification of the tropomyosin alpha-4 chain as the targeted antigen, which belongs to actin binding protein family implicated in the stabilization of the cytoskeleton actin filaments24, as well as in cell division, cytokinesis, and intracellular vesicular transport25, suggesting it as a putative target for therapy by blocking either cell division, vesicular transport and tissue remodeling.
Thiel et al.26 have described increased levels of TPM4 in tissue samples of OSCC patients. The study strategy also was different, proteins from subjects with and without OSCC were separated by 2D-PAGE, and protein patterns were identified by pairwise comparisons, and then spots were identified by MALDI-TOF mass spectrometry. Additionally, they found decreased expression of Tropomyosin 2, suggesting that tropomyosin isoforms may also present differential actions in malignant transformation. Similarly, Xiao et al. also described an up-regulation of TPM4 in tissues of oral cancer in a global proteomic analysis27, but none of the studies have investigated saliva. Moreover, TPM4 has also been found to be up-regulated in tissues, such as in breast carcinoma28,29, infiltrating ductal breast carcinoma29, colorectal neoplasia30 esophageal squamous cell carcinoma31, and scirrhous-type gastric cancer32.
Our findings present for the first time a connection between TPM4 in saliva and tissue of OSCC patients, although cross-reactions with other tumors cannot be discarded. Although none of the previous publications have shown similar results, to overcome such possibility, we should consider as an unfolding of this research also to investigate saliva samples from patients with other types of oral cancers (which represent the remaining 5% non-OSCC), and other non-oral cancers as well. Moreover, we also intend in an upcoming study to evaluate alterations in the salivary TPM4 expression in patients undergoing versus those not undergoing different cancer treatments, including chemotherapy and/or radiation. Based on the obtained results, we hypothesized that TPM4 found in saliva is probably due its release during a necrosis process, since advanced OSCC presents ulcero-proliferative growths with areas of necrotic skin33. It is also possible that the increased expression and release of TMP4 during carcinogenesis of the oral epithelium might be associated with disruption of the cytoskeleton actin filaments and/or tissue remodeling.
Briefly, we have identified TMP4 as a potential theranostic antigen in salivary samples from OSCC patients, and to our knowledge this is the first investigation that uses Phage Display with combinatorial libraries to select a functional scFv antibodies that detected a specific target in the saliva of OSCC patients, resulting in a putative marker with potential use in OSCC saliva diagnostics. Although pathological analysis of biopsies still remains the gold standard test for definitive OSCC diagnosis5, we are proposing a novel screening assay for OSCC patients through saliva, which was clinically informative (accuracy of 96.7%), faster, more convenient, easy to obtain, noninvasive and cost-effective.
Materials and Methods
Patients and sample collection
Saliva samples were obtained from 30 OSCC patients and 30 healthy donors at the Clinical Hospital of Uberlandia, Federal University of Uberlandia. Saliva was collected in the morning with a commercially available collection system (Salivette®, Sarstedt, Numbrecht, Germany). After rinsing mouths with water, two ml of saliva were obtained under a protease inhibitor solution, and samples were centrifuged at 10,000 g for 10 min at 4 °C to remove debris. The resulting supernatant was stored at −80 °C. We also obtained oral tissue from 10 patients submitted to surgical at UFU Hospital. Clinical and pathotological data were collected from patient’s charts and are listed in Table 1. The patients did not present differences in TNM classification. This study was approved by the Research Ethics Committee of the Federal University of Uberlandia (protocol number 249/2009) and informed consent was obtained from all patients. All experiments were performed in accordance with relevant guidelines and regulations.
scFvs clones anti-saliva proteins from OSCC patients
scFv clones were selected using a human immune scFv phage library19 against total proteins of saliva from OSCC patients. The library was positively selected through biopannings against total saliva proteins of OSCC patients after subtractive selection against saliva proteins from healthy subjects (negative selection) in order to eliminate major proteins with common epitopes, avoiding cross-reactivity. Briefly, two selection cycles were performed according to protocol reported elsewhere34 with modifications. Microtiter plates (Nunc MaxiSorpTM, Denmark) were coated with 10 μg of total saliva proteins diluted in 0.1 M sodium bicarbonate buffer (pH 8.6) and incubated for 18 h at 4 °C. After that, plates were blocked with 350 μL TBS/0.05% Tween 20 (TBST) and 3% BSA for 1 h at 37 °C, and washed six times with TBS. At that point, 50 μL of the library was added to the control protein well and the plate was incubated for 1 h at 37 °C. Subsequently, the unbound particles (supernatant) were transferred again to control protein well and incubated for 1 h, both steps consisted of negative selection to remove any unspecific phage. After four negative selections, the last supernatant was transferred to OSCC well and incubated for 2 h at 37 °C. The plate was washed 10 times with TBS/0.1% Tween 20, and bound phages to total proteins were eluted 100 mM glycine-HCl (pH 2.2), followed by neutralization with 16.5 μL of 2 M Tris (pH 9.1). The resulting phages from the first selection were reamplified in E. coli XL1-Blue, and a second round of selection was then performed as described above.
DNA sequencing and bioinformatics analysis
Single-strand DNA was isolated to sequencing the the heavy- and light-chain variable region genes using MegaBACE 1000 automatic sequencer (Molecular Dynamics, Sunnyvale, CA, USA). Sequencing reactions were performed with oligonucleotides for VH and VL (MMB4 and MMB5) using the DyEnamic ET Dye Terminator Cycle Sequencing Kit (GE Healthcare, USA), and the variable domain sequences and V gene families were analyzed using the Ig-BLAST and VBASE2 servers35. Sequences were further submitted to Raptor-x and PyMOL online tools to obtain and analyze the scFv 3D structure, respectively.
Soluble scFv antibody production and purification
Soluble scFv antibody was produced and purified as report elsewhere36. Briefly, clones were transformed in TOP10 E. coli and grown under agitation overnight. After incubation and centrifugation, bacteria were resuspended with proper medium overnight for antibody expression. Supernatant containing antibody was purified using Ni-affinity chromatography (HisTrapTM HP, GE Healthcare, USA), and positive fractions were pooled, desalted, lyophilized, resuspended for final quantification.
Phage antibody ELISA
Phage-ELISA was performed according the protocol performed elsewhere36. ELISA measurements were determined at 492 nm (Titertek Plus, Flow Laboratories, USA) and the cut-off values were determined by the ROC curve. Reactivity indices (RI) were obtained by dividing the ELISA readings by the cut-off value, and RI > 1 was considered positive.
Immunohistochemistry
To verify the affinity of scFv-D09 monoclonal antibody in oral squamous carcinoma tissue, immunohistochemistry was performed. For this, paraffin embedded tissue sections were deparaffinized, rehydrated and submitted to heat pretreatment in 0.1 M sodium citrate buffer for 1 h at 90 °C for antigen retrieval. Then, endogenous peroxidase activity was blocked with three 5-min washes with 3% H2O2. The sections were rinsed in distilled water and then blocked for 1 h in PBS–10% BSA. Then, approximately 4 µg/µl of the scFv-D09 were incubated in a humidifying chamber for 18 h at 4 °C. After that, slides were washed two times in PBS for 2 min each followed by incubation with HRP conjugated rat anti-HA (Roche Applied Science, Indianapolis, IN, USA) diluted (1:400) for 1 h at room temperature. After a washing step, peroxidase activity was visualized by incubation in 3, 3′- diaminobenzidine tetrahydrochloride (DAB, Sigma-Aldrich, St. Louis, MO, USA) for 5 min at room temperature and counterstained with Harris haematoxylin. Fragments of human lip well differentiated carcinoma were utilized as positive controls, and mucocele as negative control.
2-D gel electrophoresis, Mass spectrometry and bioinformatics analysis
Proteins from saliva and oral tissues were analyzedby two- dimensional gel electrophoresis (2DE)37 and two gels were performed for positive cases and negative controls. To determine the total protein concentration Bradford assay was performed38. After that, 2-D clean-up kit (GE Healthcare Life Sciences, Piscataway, NJ, USA) was used to precipitate seven hundred micrograms of protein extracts according to manufacturer’s instructions. Protein extracts were resuspended in buffer containing 6 M Urea, 2 M Thiourea, 15 mM DTT, 2% (w/v) ASB14, 0.5% IPG buffer (pH 3–10) and bromophenol blue traces and 2-D gel electrophoresis were performed according to Lazzarotto et al.39.
For Western blot analysis, the gels were transferred onto a nitrocellulose membrane (GE) and tested with scFv-D09 antibody, using the HRP-conjugated rat anti-HA (Roche Applied Science, Indianapolis, IN, USA) as secondary antibody. The identified protein spots were excised manually from the colloidal blue stained gel and subjected to trypsin digestion39. Peptide mass fingerprinting and confirmatory fragmentation analysis (MS/MS)was performed with MALDI-TOF instrument 4700 (Applied Biosystems, USA). Peak list was generated by Data Explorer v.4.5 software (Applied Biosystems, USA) using default parameters and searched with Mascot Daemon v.2.1 software (Matrix Science) against the International Protein Index (IPI) protein sequence database (IPI; version 3.12). Only significant hits (P > 0.05) were accepted, according to the MASCOT probability method. Then, we used the molecular mass and pI values obtained in 2D gel electrophoresis to confirm and identify the target antigen. Furthermore, the highest similar protein sequence was submitted to bioinformatics online tools. To obtain the 3D structure of the protein, Raptor-x was used. For identification of potential binding regions of the protein and scFv it docking was performed. The regions of scFv-protein structure were selected and analyzed by PyMOL.
Statistical analysis
GraphPad Prism 6.0 (GraphPad Soſtware Inc., San Diego, USA) was used to perform statistical analyses. Receiver operating characteristics (ROC) curves were constructed to define the best cut-off value and diagnostic parameters: sensitivity (Se), specificity (Sp), likelihood ratio (LR+) and respective area under the curve (AUC) with 95% confidence interval (CI). The Mann–Whitney test (nonparametric analyses) was used to determine differences between Control and cancer group in ELISA. Probability below 0.05 was considered significant.
References
Bray, F., Ferlay, J. & Soerjomataram, I. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. 394–424, https://doi.org/10.3322/caac.21492 (2018).
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer Statistics, 2018. 68, 7–30 (2018).
INCA. Estimativa 2018: incidência de câncer no Brasil. (Ministry of Health – Brazilian National Cancer Institute José Alencar Gomes da Silva (INCA), 2018).
Mishra, R. Biomarkers of oral premalignant epithelial lesions for clinical application. Oral Oncol. 48, 578–584 (2012).
Carreras-Torras, C. & Gay-Escoda, C. Techniques for early diagnosis of oral squamous cell carcinoma: Systematic review. Med. Oral Patol. Oral y Cir. Bucal e305–e315, https://doi.org/10.4317/medoral.20347 (2015).
Yu, J.-S. et al. Saliva protein biomarkers to detect oral squamous cell carcinoma in a high-risk population in Taiwan. Proc. Natl. Acad. Sci. 113, 11549–11554 (2016).
Gallo, C. et al. Potential Salivary Proteomic Markers of Oral Squamous Cell Carcinoma. Cancer Genomics Proteomics 13, 55–61 (2016).
Malamud, D. Saliva as a Diagnostic Fluid. Dent. Clin. North Am. 55, 159–178 (2011).
Bonne, N. J. & Wong, D. T. Salivary biomarker development using genomic, proteomic and metabolomic approaches. Genome Med. 4, 82 (2012).
Juretić, M. et al. Salivary levels of TNF-α and IL-6 in patients with oral premalignant and malignant lesions. Folia Biol. (Praha). 59, 99–102 (2013).
St. John, M. A. R. et al. Interleukin 6 and Interleukin 8 as Potential Biomarkers for Oral Cavity and Oropharyngeal Squamous Cell Carcinoma. Arch. Otolaryngol. Neck Surg. 130, 929 (2004).
Zanotti, L. et al. Epidermal growth factor receptor detection in serum and saliva as a diagnostic and prognostic tool in oral cancer. Laryngoscope 127, E408–E414 (2017).
Gaba, F. I., Sheth, C. C. & Veses, V. Salivary biomarkers and their efficacies as diagnostic tools for Oral Squamous Cell Carcinoma: Systematic review and meta-analysis. J. Oral Pathol. Med., https://doi.org/10.1111/jop.12791 (2018).
Rajkumar, K. et al. Salivary and serum level of CYFRA 21-1 in oral precancer and oral squamous cell carcinoma. Oral Dis. 21, 90–96 (2015).
Hu, S. et al. Salivary Proteomics for Oral Cancer Biomarker Discovery. Clin. Cancer Res. 14, 6246–6252 (2008).
Mizukawa, N. et al. Defensin-1, a peptide detected in the saliva of oral squamous cell carcinoma patients. Anticancer Res. 18, 4645–9
Dalli, J., Montero-Melendez, T., McArthur, S. & Perretti, M. Annexin A1 N-Terminal Derived Peptide Ac2-26 Exerts Chemokinetic Effects on Human Neutrophils. Front. Pharmacol. 3, 28 (2012).
Araújo, T. G. et al. A novel highly reactive Fab antibody for breast cancer tissue diagnostics and staging also discriminates a subset of good prognostic triple-negative breast cancers. Cancer Lett. 343, 275–285 (2014).
Carneiro, A. P. et al. A putative OTU domain-containing protein 1 deubiquitinating enzyme is differentially expressed in thyroid cancer and identifies less-aggressive tumours. Br. J. Cancer 111, 551–558 (2014).
Reis, C. F. et al. An antibody-like peptide that recognizes malignancy among thyroid nodules. Cancer Lett. 335, 306–313 (2013).
McCafferty, J., Griffiths, A. D., Winter, G. & Chiswell, D. J. Phage antibodies: filamentous phage displaying antibody variable domains. Nature 348, 552–554 (1990).
Krapfenbauer, K., Drucker, E. & Thurnher, D. Identification of tumour-related proteins as potential screening markers by proteome analysis—protein profiles of human saliva as a predictive and prognostic tool. EPMA J. 5, 20 (2014).
Sivadasan, P. et al. Human salivary proteome — a resource of potential biomarkers for oral cancer. J. Proteomics 127, 89–95 (2015).
Wen, K.-K., Kuang, B. & Rubenstein, P. A. Tropomyosin-dependent Filament Formation by a Polymerization-defective Mutant Yeast Actin (V266G,L267G). J. Biol. Chem. 275, 40594–40600 (2000).
Gunning, P., O’neill, G. & Hardeman, E. Tropomyosin-Based Regulation of the Actin Cytoskeleton in Time and Space. Physiol. Rev. 88, 1–35 (2008).
Thiel, U. J. E. et al. Analysis of differentially expressed proteins in oral squamous cell carcinoma by MALDI-TOF MS. J. Oral Pathol. Med. 40, 369–379 (2011).
Xiao, H. et al. Quantitative proteomic analysis of microdissected oral epithelium for cancer biomarker discovery. Oral Oncol. 51, 1011–1019 (2015).
Li, D.-Q. et al. Identification of breast cancer metastasis-associated proteins in an isogenic tumor metastasis model using two-dimensional gel electrophoresis and liquid chromatography-ion trap-mass spectrometry. Proteomics 6, 3352–3368 (2006).
Kabbage, M. et al. Tropomyosin-4 correlates with higher SBR grades and tubular differentiation in infiltrating ductal breast carcinomas: an immunohistochemical and proteomics-based study. Tumor Biol. 34, 3593–3602 (2013).
Polley, A. C. J. et al. Proteomic Analysis Reveals Field-Wide Changes in Protein Expression in the Morphologically Normal Mucosa of Patients with Colorectal Neoplasia. Cancer Res. 66, 6553–6562 (2006).
Harada, T. et al. Expression of tropomyosin alpha 4 chain is increased in esophageal squamous cell carcinoma as evidenced by proteomic profiling by two-dimensional electrophoresis and liquid chromatography-mass spectrometry/mass spectrometry. PROTEOMICS – Clin. Appl. 1, 215–223 (2007).
Kuramitsu, Y. et al. Proteomic differential display analysis shows up-regulation of 14-3-3 sigma protein in human scirrhous-type gastric carcinoma cells. Anticancer Res. 30, 4459–65 (2010).
Barnes, L. E. J., Reichart, P. & Sidransky, D. World Health Organization Classification of Tumours: Pathology & Genetics Head and Neck Tumours IARC (2005).
Carlos F Barbas III, D R Burton, G. J. S. Phage display: A laboratory manual.
Retter, I. VBASE2, an integrative V gene database. Nucleic Acids Res. 33, D671–D674 (2004).
Levenhagen, M. A. et al. Structural and functional characterization of a novel scFv anti-HSP60 of Strongyloides sp. 1–10, https://doi.org/10.1038/srep10447.
Roessler, M. et al. Identification of PSME3 as a Novel Serum Tumor Marker for Colorectal Cancer by Combining Two-dimensional Polyacrylamide Gel Electrophoresis with a Strictly Mass Spectrometry-based Approach for Data Analysis. Mol. Cell. Proteomics 5, 2092–2101 (2006).
Bradford, M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–54 (1976).
C, L.-S. et al. Similar proteomic profiles of human mesenchymal stromal cells from different donors. Cytotherapy 11, 268–77 (2009).
Acknowledgements
The authors thank the Brazilian funding agencies, CNPq, CAPES and FAPEMIG, for providing financial support to the National Institute of Science and Technology in Theranostics and Nanobiotechnology – INCT-TeraNano (CNPq/CAPES/FAPEMIG, Grant numbers CNPq-465669/2014-0 and FAPEMIG-CBB-APQ-03613-17).
Author information
Authors and Affiliations
Contributions
P.C.B.F. and L.R.G. designed the study; P.C.B.F., A.P.C., R.B., R.N., P.S.S. and D.F. performed experiments and analyzed data; A.M.L. and E.A. contributed with new reagents/analytic tools (immunohistochemistry and bidimensional electrophoresis. S.J.S. and A.M.L. made patient’s recruitment and clinical evaluations; P.C.B.F., A.P.C. and L.R.G. wrote the paper. All authors discussed the results and implications and contributed to the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors L.R.G., P.C.B.F., P.S.S., R.N. and A.P.C. are co-authors of the patent application BR1020170211320, filed on Oct. 2nd, 2017, where part of the findings present in the manuscript are claimed.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Faria, P.C.B., Carneiro, A.P., Binato, R. et al. Upregulation of tropomyosin alpha-4 chain in patients’ saliva with oral squamous cell carcinoma as demonstrated by Phage display. Sci Rep 9, 18399 (2019). https://doi.org/10.1038/s41598-019-54686-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-54686-x