Abstract
The SARS-CoV-2 non-structural protein 1 (Nsp1), also referred to as the host shutoff factor, suppresses host innate immune functions. By combining cryo-electron microscopy and biochemistry, we show that SARS-CoV-2 Nsp1 binds to the human 40S subunit in ribosomal complexes, including the 43S pre-initiation complex and the non-translating 80S ribosome. The protein inserts its C-terminal domain into the mRNA channel, where it interferes with mRNA binding. We observe translation inhibition in the presence of Nsp1 in an in vitro translation system and in human cells. Based on the high-resolution structure of the 40S–Nsp1 complex, we identify residues of Nsp1 crucial for mediating translation inhibition. We further show that the full-length 5′ untranslated region of the genomic viral mRNA stimulates translation in vitro, suggesting that SARS-CoV-2 combines global inhibition of translation by Nsp1 with efficient translation of the viral mRNA to allow expression of viral genes.
Similar content being viewed by others
Main
SARS-CoV-2 is the causing agent of the COVID-19 pandemic and belongs to the genus of beta-coronaviruses, with enveloped, positive sense and single-stranded genomic RNA1. On entering host cells, the viral genomic RNA is translated by the cellular protein synthesis machinery to produce a set of non-structural proteins (NSPs)2. NSPs render the cellular conditions favorable for viral infection and viral mRNA synthesis3. Coronaviruses have evolved specialized mechanisms to hijack the host gene expression machinery and employ cellular resources to regulate viral protein production. Such mechanisms are common for many viruses and include inhibition of host protein synthesis and endonucleolytic cleavage of host messenger RNAs (mRNAs)4,5. In cells infected with the closely related SARS-CoV, one of the most enigmatic viral proteins is the host shutoff factor, Nsp1. Nsp1 is encoded by the gene closest to the 5′ end of the viral genome and is among the first proteins to be expressed after cell entry and infection to repress multiple steps of host protein expression6,7,8,9. Initial structural characterization of the isolated SARS-CoV Nsp1 protein revealed the structure of its N-terminal domain, whereas its C-terminal region was flexibly disordered10. Furthermore, it was shown that SARS-CoV Nsp1 suppresses host innate immune functions, mainly by targeting type I interferon expression and antiviral signaling pathways11. Taken together, Nsp1 serves as a potential virulence factor for coronaviruses and represents an attractive target for live attenuated vaccine development12,13.
To provide molecular insights into the mechanism of SARS-CoV-2 Nsp1-mediated translation inhibition, we solved the structures of ribosomal complexes isolated from HEK293 lysates supplemented with recombinant purified Nsp1 as well as of an in vitro reconstituted 40S–Nsp1 complex using cryo-EM. We complement our findings by reporting in vitro and in vivo translation inhibition in the presence of Nsp1 that is relieved after mutating key interacting residues. Furthermore, we show that the translation output of reporters containing full-length viral 5′ untranslated regions (UTRs) is significantly enhanced, which could explain how Nsp1 inhibits global translation while still translating sufficient amounts of viral mRNAs.
Results
C-terminal domain of SARS-CoV-2 Nsp1 binds to the mRNA entry channel
To elucidate the mechanism of how Nsp1 inhibits translation, we aimed to identify the structures of potential ribosomal complexes as binding targets (Fig. 1). Previously, it has been suggested that Nsp1 mainly targets the ribosome at the translation initiation step9. We thus treated lysed HEK293E cells with bacterially expressed and purified Nsp1 and loaded the cleared lysate on a sucrose gradient. Fractions containing ribosomal particles were then analyzed for the presence of Nsp1. Interestingly, Nsp1 not only co-migrated with 40S particles, but also with 80S ribosomal complexes (Fig. 1c), suggesting that it interacts with a range of different ribosomal states. We then pooled all sucrose gradient fractions containing ribosomal complexes and investigated them using cryo-EM (Table 1). This analysis revealed a 43S pre-initiation complex (PIC) encompassing the initiation factor eIF3 core, eIF1, eIF1A, the ternary complex comprising eIF2 and initiator tRNAi, as well as an additional density in the mRNA entrance channel that could not be assigned to mRNA (Fig. 1a and Extended Data Fig. 1). The same density was also observed in a translationally inactive 80S complex in which only an E-site tRNA was bound, but no mRNA (Fig. 1b and Extended Data Fig. 1). These findings suggest that the extra density can be assigned to Nsp1 that competes with mRNA for ribosome binding to inhibit translation. To unambiguously attribute the observed additional density in the 43S PIC and 80S complex to Nsp1, we assessed whether Nsp1 binds purified ribosomal 40S subunits alone. In vitro binding assays using sucrose density centrifugation showed that Nsp1 associates with 40S ribosomal subunits, because it co-pelleted with the 40S (Fig. 1d). However, Nsp1 did not interact with 60S subunits, suggesting that the interaction with 40S subunits is specific. Based on these results we assembled a 40S–Nsp1 complex in vitro and determined its structure at 2.8-Å resolution using cryo-EM (Extended Data Fig. 2). The molecular details revealed by these maps allowed us to identify the density as the C-terminal region of Nsp1 and build an atomic model (Table 1). Docking of the model into the maps of the 43S PIC and 80S obtained from the HEK293E cell lysates clearly showed that the C terminus of Nsp1 is also associated with the 40S ribosomal subunits in these complexes (Fig. 1a,b and Extended Data Fig. 3). As observed in the high-resolution structure of the 40S–Nsp1 complex, the C-terminal part of Nsp1 in the mRNA entrance channel folds into two helices that interact with h18 of the 18S rRNA as well as proteins uS3 in the head and uS5 and eS30 in the body, respectively (Figs. 1e and 2a). In all complexes, Nsp1 binds in the mRNA entrance channel on the 40S subunit, where it would partially overlap with the fully accommodated mRNA. Consequently, mRNA was not observed due to Nsp1 binding.
a, Views of Nsp1 (red) binding to a 43S PIC containing the core of initiation factor eIF3 (cyan), eIF1 (blue) and the eIF2–tRNA ternary complex (magenta). b, Overview of Nsp1 (red) binding to a translationally inactive 80S complex. c, Sucrose gradient fractionation of HEK lysate supplemented with Nsp1. Nsp1 co-migrates with 40S and 80S ribosomal particles in a 15–45% (wt/vol) sucrose gradient. His6-tagged Nsp1 is visualized by western blot using an α-His antibody (fractions 1–20 and loading control (c), with 0.2 μg of purified His6-Nsp1 applied), while the rRNA content in corresponding fractions is monitored on an agarose gel. All samples for the western blot are derived from the same experiment and the blots were processed in parallel. d, In the in vitro binding assay, wild-type (WT) Nsp1 was added to 40S and 60S ribosomal subunits and loaded on a 30% (wt/vol) sucrose cushion. Unbound proteins remained in the supernatant (SN), while bound Nsp1 co-pelleted with 40S (P). e, Overview of Nsp1 binding to the small ribosomal subunit. Nsp1 (red) binds close to the mRNA entry site and contacts uS3 (blue) from the ribosomal 40S head as well as uS5 (green), the C terminus of eS30 (orange) and h18 of the 18S rRNA (gray) of the 40S body. A magnified view of the Nsp1 binding area is shown in the box. Uncropped gel images for c and d are available online as source data.
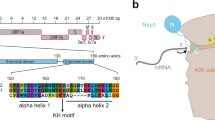
a, Overview of Nsp1 binding to uS5 and the 18S rRNA. Nsp1 specifically interacts with uS5 through a hydrophobic patch and is tightly anchored to helix h18 of the 40S rRNA with a set of positively charged residues. b–d, Detailed views of the specific interaction areas indicated in a. The 2.8-Å experimental EM densities are shown as dark blue mesh and are contoured at 7.3σ. Dashed lines indicate contacts between positively charged Nsp1 residues and the rRNA backbone within hydrogen-bonding distance (<3.8 Å). Residues mutated in this study are highlighted in orange (KH), green (YF) and blue (RR). e, Alignment of the Nsp1 C termini from human SARS-CoV, SARS-CoV-2 and closely related bat coronaviruses. Residues that mediate the interaction with the 40S mRNA entry site are highly conserved. Pairs of amino acids mutated for functional studies are highlighted. An overview of the Nsp1 domain arrangement, including the flexible linker, is depicted in the schematic.
Molecular details of translation inhibition by Nsp1
The high-resolution reconstruction also revealed the network of molecular interactions between Nsp1 and the 40S subunit. The first C-terminal helix (residues 153–160) interacts with uS5 and uS3 through multiple hydrophobic side chains such as Y154, F157 and the following residue W161 (Fig. 2b). The two helices are connected by a short loop containing the KH motif that establishes stacking interactions with helix h18 of the 18S rRNA through U607 and U630 as well as backbone binding (Fig. 2c). The second helix (residues 166–178), localized in the proximity of the eS30 C terminus, interacts with the phosphate backbone of h18 via the two conserved arginines R171 and R175 (Fig. 2d).
An additional weak density was observed at the head of the small subunit in the proximity of eS10 between h16 and uS3. This may correspond to the flexibly disposed N-terminal domain of Nsp1 considering the 20-amino acid unstructured linker between the N- and C-terminal domains (Fig. 2e and Supplementary Fig. 1). However, we cannot exclude that this density corresponds to unassigned ribosomal protein segments in the vicinity, as the C-terminal 65 amino acids of eS10 (head) or the N-terminal 60 amino acids of uS5 (body) could become better ordered in the context of Nsp1 binding. Thus, Nsp1 is tightly bound to the 40S subunit through anchoring of its C-terminal helices to the mRNA channel, while the N-terminal domain can sample space within a radius of ~60 Å from its attachment point.
Nsp1 WT efficiently inhibits translation, while Nsp1 mutants lose their ribosome binding and inhibition capability
We further investigated how wild-type (WT) Nsp1 affects translation of a Renilla Luciferase-encoding reporter mRNA (RLuc) in an S3 HeLa lysate in vitro translation system14. WT Nsp1 was recombinantly expressed and purified and its effect on translation was tested by adding increasing amounts of the protein to HeLa cell lysates containing a capped and polyadenylated RLuc mRNA control transcript (Fig. 3). We observed a concentration-dependent inhibition of translation where almost full inhibition was reached at 1 µM Nsp1 (Fig. 3b,d).
a, The C-terminal domain of Nsp1 binds to the mRNA entry site of 40S (red), while the N-terminal domain (gray) is flexibly disposed. Mutants in helix 2 (Y154A/F157A in green), in the short connecting loop (K164A/H165A in orange), in helix 3 (R171E/R175E in blue) and a truncation of the C-terminal domain were generated. In the Trx1-Linker-Cter Nsp1 construct, the N-terminal domain was replaced by Escherichia coli Trx1 (purple). b, Relative RLuc activity measurements of in vitro translation reactions normalized to the reaction in the absence of Nsp1 (0 μM). Data are presented as mean values ± standard deviations of three biological replicates (sets of translation reactions) averaged after three measurements; mean values of each biological replicate are indicated by dots. One-way analysis of variance (ANOVA) analysis is compared to the 0 μM Nsp1 condition. NS, not statistically significant (>0.05); **P < 0.01, ****P < 0.0001. c, In the in vitro binding assay, Nsp1 mutants were added to 40S and 60S ribosomal subunits and loaded on a 30% (wt/vol) sucrose cushion. SN, supernatant; P, pellet. d, Scheme of the in vitro synthesized capped (black dot) and polyadenylated reporter mRNAs coding for Renilla Luciferase (hRLuc). e, RLuc activity measurements of in vitro translation reactions normalized to the readout of RLuc reporter mRNA. Bars, mean values and dots are defined in b. P value of an unpaired t-test, 0.014. f, Titration of WT Nsp1 against RLuc and FL-RLuc reporter mRNAs. Relative RLuc activities were normalized to the untreated sample (0 μM). Bars, mean values and dots are as defined in b. One-way ANOVA analysis is compared to the 0 μM Nsp1 condition and the P values were all <0.0001. ORF, open reading frame. Source data for b, e and f and uncropped gel images for c are available online.
To dissect the contributions of the observed interactions between the 40S and Nsp1 on the inhibition of translation, we used our structural information to design several mutants targeting key amino acids in helix 1 (double mutant Y154A/F157A), the KH motif (double mutant K164A/H165A) and in helix 2 (double mutant R171E/R175E), as summarized in Fig. 3a. We also rationalized our mutations based on the high conservation of the Nsp1 C terminus between SARS-CoV-2, SARS-CoV and closely related bat Coronaviridae, with sequence identities above 85% for the ORF1ab (encoding for polyproteins pp1a and pp1ab) and key amino acids highly conserved (Fig. 2e and Supplementary Fig. 1)15. Furthermore, the KH mutant has been described to abolish interaction with 40S in SARS-CoV11. In contrast to the WT protein, the three mutants did not affect translation of the RLuc control mRNA, even at concentrations of 6 µM (Fig. 3b). Consistently, when deleting the entire C-terminal domain (Nter-Linker Nsp1), translation was no longer inhibited (Fig. 3a,b). All described mutants also lost their ability to bind 40S ribosomal subunits (Fig. 3c and Extended Data Fig. 4a), indicating that the interaction of the C-terminal domain with the 40S ribosomal subunit is primarily responsible for the observed translation inhibition. These results also agree with our structural findings that the C-terminal domain of Nsp1 is responsible for specific contacts with the ribosome, whereas the N-terminal domain is flexibly disposed. Interestingly, this inhibition mechanism may be unique to SARS-CoV-2 and closely related beta-coronaviruses, because the C-terminal region of Nsp1 is shorter in alpha-coronaviruses and is not highly conserved amongst other beta-coronaviruses, including MERS-CoV. This is consistent with the observation that MERS-CoV Nsp1 does not bind the ribosome16.
To investigate whether Nsp1 can inhibit translation in vivo, we transiently expressed FLAG-tagged WT Nsp1 or the KH mutant in HeLa cells and assessed the effect on global translation using a puromycin incorporation assay. It is noteworthy that transfection of equal amounts of plasmid DNA resulted in much lower WT Nsp1 protein levels than with the KH mutant. This possibly indicates an autoregulatory feedback loop by which Nsp1 starts to inhibit its own synthesis above a certain intracellular concentration (Fig. 4a). Therefore, transfected plasmid DNA amounts were adjusted for the puromycin incorporation assay to achieve similar expression levels of WT and KH mutant Nsp1. Compared to untransfected cells, WT Nsp1 caused a reduction of puromycin incorporation and translation efficiency to ~50% (Fig. 4b–d). Similar levels of the KH mutant did not affect the translation efficiency, which corroborates the results of the in vitro translation assays and further highlights the importance of the KH motif for ribosome binding and translation inhibition.
a, Western blot analysis of HeLa cells expressing FLAG-Nsp1. HeLa cells were transfected with equal amounts of plasmid encoding either wild-type FLAG-tagged Nsp1 (WT) or FLAG-tagged Nsp1 with a mutated KH motif (KH). Cells transfected with empty vector served as the control (Ctrl). Whole-cell lysates were analyzed by western blotting using anti-FLAG and anti-GAPDH antibodies. b,c, Results of a puromycin incorporation assay in the presence of Nsp1. WT or KH FLAG-Nsp1 were transiently expressed in HeLa cells. Plasmid amounts were adjusted to achieve similar protein levels. As a control, one culture was incubated with cycloheximide (CHX). Before collection, nascent proteins were labeled with a puromycin pulse. Cell lysates were prepared and the Nsp1 expression (b, anti-FLAG) and incorporated puromycin (c, anti-puromycin) were analyzed by western blotting. GAPDH served as loading control. d, Quantification of puromycin incorporation. The intensity of each lane on the anti-puromycin stained blot (c) was measured and normalized to the intensity of the loading control. Bars represent mean values of three independent experiments (indicated by single dots) relative to the control transfection ± standard deviation (one-way ANOVA test P values: Ctrl versus WT, 0.0006; Ctrl versus KH, 0.9211; Ctrl versus CHX, <0.0001). Uncropped gel images for a–c and source data for d are available online.
Given that we were not able to place the N-terminal domain of Nsp1 with confidence in our cryo-EM maps, we attempted to further dissect its role by replacing the domain with the soluble domain of E. coli thioredoxin 1 (Trx1), which has no known affinity for the ribosome. This Nsp1 construct was still able to bind 40S ribosomal subunits in our sucrose pelleting binding assay and efficiently inhibited translation in vitro (Fig. 3a,b and Extended Data Fig. 4b). These results indicate that the C-terminal domain is necessary and sufficient for translation inhibition. Although the N-terminal domain was suggested to play a role in the regulation of cellular mRNA stability10 or in suppressing host innate immune functions11, we show that it does not play an important role in the inhibition of translation.
Because translation of viral mRNA competes with translation of cellular mRNAs, the inhibitory effects of Nsp1 in the context of special features of the SARS-CoV-2 genomic RNA need to be considered. We thus investigated the differences in the translation of reporter mRNAs with viral versus cellular 5′ UTRs and the relative inhibitory effect of Nsp1. Using the in vitro translation system described above, we compared the translation efficiency of capped and polyadenylated RLuc reporters harboring the full-length 5′ UTR of the SARS-CoV-2 genomic RNA (FL-RLuc) with the translation of equimolar amounts of a native RLuc reporter (Fig. 3d and Extended Data Fig. 5b). We observed a significant fivefold increase in translation when the reporter mRNA included the viral 5′ UTR, indicating that the viral mRNA is more efficiently translated than host mRNAs (Fig. 3e). Nevertheless, titration of WT Nsp1 inhibited translation of both mRNAs, FL-RLuc and native RLuc, to the same extent (Fig. 3f).
Discussion
In this work we show that SARS-CoV-2 Nsp1 causes translation inhibition by sterically occluding the entrance region of the mRNA channel in the free 40S subunits, the 43S pre-initiation complex and in empty, non-translating 80S ribosomes (Fig. 5a,b). It was previously biochemically shown for the SARS-causing virus SARS-CoV8 that Nsp1 inhibits translation, which suggests that this role of Nsp1 is conserved among most beta-coronaviruses. A similar mode of ribosome binding is also employed by some cellular factors such as SERPINE mRNA-binding protein 1 (SERBP1), which binds to the mRNA channel spanning from the mRNA channel entrance across the A and P sites to prevent mRNA binding and to sequester 80S ribosomes17,18. Interestingly, in our experiments using HEK lysates we also observed a class of the 80S ribosomes that are associated with SERBP1 and elongation factor eEF2 (Extended Data Fig. 1).
a, Superposition of canonically bound mRNA (green), A- (blue) and P-site (purple) tRNAs (PDB 6HCJ) reveals that Nsp1 (red) prevents classical binding of the mRNA at the entry site due to blockage. b, Nsp1 binds via its C terminus in proximity to the 40S mRNA entry site. Because of the flexible linker, the N-terminal domain can sample an area of ~60 Å around its attachment point (circle). c, Model for translation inhibition by Nsp1. Following viral infection and translation of viral genomic mRNA (red), Nsp1 acts as a translation inhibitor, reducing the pool of ribosomes that can engage in translation. Under such ribosome-limiting conditions, translation of viral mRNAs will be favored over translation from less efficient cellular 5′ UTRs (blue).
Our experiments show that the 5′ UTR of SARS-CoV-2 efficiently promotes translation initiation; it remains unclear, however, how this efficiency is achieved. This region of viral RNA has a complex secondary structure and is conserved in most coronaviruses (Extended Data Fig. 5a), where various stem loops have been described to play functional roles in viral replication, including discontinuous transcription and translation19,20,21. This region might also enhance initiation of translation through interactions with the 40S subunit, translation initiation factors, or it might speed up the transition from initiation to elongation, thus leading to higher ribosome loading.
Our data demonstrate that Nsp1 inhibits translation of both a native and a viral 5′ UTR-containing reporter mRNA. This implies that viral mRNAs do not evade translation inhibition in the context of the 5′ UTR sequence. However, the observation that mRNAs with viral 5′ UTR are translated more efficiently provides a possible explanation of how the translation machinery is hijacked by the virus. First, Nsp1 acts as a strong inhibitor of translation, tightly binding ribosomes and reducing the pool of available ribosomes that can engage in translation. Under such ribosome-limiting conditions, translation from less abundant but more efficient mRNAs is likely to be favored over the mRNAs with less efficient 5′ UTRs (Fig. 5c). Therefore, the combination of an Nsp1-mediated general translation inhibition and a highly efficient translation of viral transcripts may explain how viral infected cells switch their translation from host cell mRNAs towards viral mRNAs. Considering that Nsp1 inhibits its own translation, the virus may tune the cellular levels of Nsp1 below the concentrations that are needed to abolish viral mRNA translation, but enough to inhibit translation initiation from less efficiently recruited cellular mRNAs. Through this mechanism, we propose that Nsp1 would be able to considerably reduce global cellular translation, while the remaining ribosomes would still be able to translate viral mRNAs with high efficiency. During the course of viral infection, the effect of viral mRNAs on shifting protein synthesis machinery towards the production of viral proteins would be increasingly strong because their levels are known to increase to 50% of total cellular RNAs22.
A related paper was recently published describing how, upon overexpression of a tagged Nsp1 in human cells, Nsp1 binds to various ribosomal complexes, including translationally inactive ribosomes, pre-40S-like complexes and 43S pre-initiation complexes, and showing that Nsp1 blocks innate immune responses23 as described previously for SARS-CoV11. Although we followed a different experimental strategy to generate the complexes in human cell lysates using an untagged protein added at a defined concentration, the results reported in that study are fully consistent and complementary to our findings.
The identification of the C-terminal region of Nsp1 as the key domain for ribosome interactions that are essential for controlling cellular response to viral infections will be helpful in designing attenuated strains of SARS-CoV-2 for vaccine development. Additionally, molecular insights into the Nsp1-mediated translation inhibition could prove valuable for the design and selection of potential Nsp1 small-molecule inhibitors24. Our results provide an excellent basis for structure-based experiments aimed at investigating Nsp1 function in vivo by using viral model systems.
Methods
Cell lines
For the puromycilation experiments, HeLa (ATCC; CCL2) cells were used and tested negative for mycoplasma. HeLa cells were cultured in DMEM supplemented with 10% FCS supplemented with 100 UI ml−1 penicillin and 100 μg ml−1 streptomycin (DMEM+/+) at 37 °C, 5% CO2. For generation of the HEK lysate, 293 c18 (ATCC CRL-10852), a human HEK293-EBNA cell line, was used, and cells were tested negative for mycoplasma.
Cloning, expression and purification of Nsp1 in E. coli
Plasmids encoding the WT Nsp1 sequence and the N-terminal domain replaced by the soluble domain of E. coli thioredoxin were ordered from GenScript Biotech. Plasmids encoding Nsp1 protein mutants were generated by site-directed mutagenesis using the primers 5′-GTT CAC GGG TCA CAC CGC TGC TAG CTG CGG TAT TCC AAT TTT CCT GAA AAT-3′, 5′-CGA GTT AGC CAC CAT TCA GTT CCT CCA TCA GTT CCT CGG TCA CAC CGC TGC TAT GTT TG-3′ and 5′-TTT GGT ATT CCA ATT TTC CTG AGC ATC TTC AGC CGG ATC GGT GCC CAG TTC ATC G-3′ to yield KHAA, RREE and YFAA mutants, respectively. Nsp1 (WT and mutants) carrying an N-terminal His6-tag followed by a tobacco etch virus (TEV) cleavage site was expressed from a pET24a vector. The plasmid was transformed into E. coli BL21-CodonPlus (DE3)-RIPL and cells were grown in 2× YT medium at 30 °C. At an optical density at 600 nm (OD600) of 0.8, cultures were shifted to 18 °C and induced with IPTG added to a final concentration of 0.5 mM. After 16 h, cells were collected by centrifugation, resuspended in lysis buffer (50 mM HEPES-KOH pH 7.6, 500 mM KCl, 5 mM MgCl2, 40 mM imidazole, 10% (wt/vol) glycerol, 0.5 mM TCEP and protease inhibitors) and lysed using a cell disrupter (Constant Systems). The lysate was cleared by centrifugation for 45 min at 48.000g and loaded onto a HisTrap FF 5-ml column (GE Healthcare). Eluted proteins were incubated with TEV protease at 4 °C overnight and both the His6-tag, uncleaved Nsp1 and the His6-tagged TEV protease were removed on the HisTrap FF 5-ml column. The sample was further purified via size-exclusion chromatography on a HiLoad 16/60 Superdex75 system (GE Healthcare), buffer exchanging the sample to the storage buffer (40 mM HEPES-KOH pH 7.6, 200 mM KCl, 40 mM MgCl2, 10% (wt/vol) glycerol, 1 mM TCEP). Fractions containing Nsp1 were pooled, concentrated in an Amicon Ultra-15 centrifugal filter (10-kDa molecular weight cutoff (MWCO)), flash-frozen in liquid nitrogen and stored until further use at −80 °C.
Preparation of human ribosomal subunits
Human ribosomal subunits were purified as described in ref. 25 and the final samples were flash-frozen in liquid nitrogen at a concentration of 1 mg ml−1 (OD600 of 10) and stored at −80 °C.
Sucrose pelleting binding assay
To verify Nsp1–40S complex formation, we performed binding assays using sucrose density centrifugation. Thawed human 40S and 60S ribosomal subunits were adjusted to a final concentration of 0.3 μM in a 100 μl reaction (complex binding buffer: 20 mM HEPES pH 7.6, 5 mM MgCl2, 100 mM KCl, 2 mM DTT) and mixed with a 5× molar excess of His6-Nsp1 WT/mutant. The assembled complexes were incubated for 5 min at 30 °C and for 10 min on ice before they were loaded on a 30% (wt/vol) sucrose cushion in TLA-100 tubes (Beckman Coulter). The sucrose cushions were centrifuged for 2 h at 390,880g and 4 °C. After removing the supernatant, the pellet was resuspended in 20 μl of complex binding buffer. Samples were analyzed on bleach agarose gels (0.06% bleach, 1% (wt/vol) agarose) for visualization of the RNA and on western blot (anti-His antibody, Clontech) for visualization of His6-tagged Nsp1.
Preparation of viral 5′-UTR mRNA and reporter RLuc mRNA
Plasmids
MS2-containing mRNA reporters (p200-6xMS2) were derived from the pCRII-hRLuc-200bp 3′ UTR construct as described previously14. Amplification of the vector using the primers 5′-AAT AAG AGC TCC TGC CTC GAG CTT CCT CATC-3′ and 5′-AAT AAC ATA TGG TGA TGC TAT TGC TTT ATT TGT AAC-3′ was followed by restriction digestion with SacI and NdeI and ligation with a 6xMS2-containing insert that was PCR-amplified using the primers 5′-AAT AAC ATA TGG TTC CCT AAG TCC AAC TAC CAA A-3′ and 5′-AAT AAA GAG CTC CCA GAG GTT GAT TGT CGA CC-3′ and that had been treated with the same enzymes. The 265-nt-long 5′ UTR of SARS-CoV genomic mRNA sequence was subcloned to replace the RLuc 5′ UTR by fusion PCR using primers TCTGCAGAATTCGCCCTTCATG and GCCCTATAGTGAGTCGTATTACAATTCACT for vector amplification and the pair GACTCACTATAGGGCATTAAAGGTTTATACCTTCCCAGGTAACAAAC and GGCGAATTCTGCAGACTTACCTTTCGGTCACACCCG for amplification of the 5′ UTR fragment using 5′ UTR-eGFP cloned in pUC19 vector as a template, which was designed to possess the SARS-CoV-2 5′ UTR sequence in front of the eGFP coding sequence.
In vitro transcription of reporter mRNAs
Preparation of in vitro transcribed mRNAs was performed as described in ref. 14; namely, linearized pCRII vectors encoding the desired reporter mRNA downstream of a T7 promoter were mixed to yield an in vitro transcription reaction in 1× transcription buffer (Thermo Fisher Scientific) at a final concentration of 20–30 ng µl−1. This mixture further contained each ribonucleotide at 1 mM (rNTPs, Thermo Fisher Scientific), 1 U µl−1 murine RNase inhibitor (Vazyme), 0.001 U µl−1 pyrophosphatase (Thermo Fisher Scientific) and 5% (vol/vol) T7-RNA-polymerase (custom-made). The reaction was incubated at 37 °C for 1 h and then an equal quantity of T7-RNA polymerase was added for another 30 min. The mixture was then supplemented with TURBO DNase (Thermo Fisher Scientific) to a final concentration of 0.14 U µl−1 and incubated at 37 °C for 30 min. The transcribed mRNA was purified from the reaction using an acidic phenol-chloroform-isoamylalcohol (PCI). The product was dissolved in disodium citrate buffer, pH 6.5, and quality was assessed by agarose gel electrophoresis.
Before capping, the RNA was incubated at 65 °C for 5 min and supplemented accordingly to yield a reaction consisting of 300 ng µl−1 RNA, 0.5 mM guanosine triphosphate (GTP, New England Biolabs), 0.1 mM S-adenosylmethionine (SAM, New England Biolabs), 1 U µl−1 murine RNase inhibitor (Vazyme), 0.5 U µl−1 vaccinia capping enzyme (New England Biolabs) in 1× capping buffer (New England Biolabs). The capping reaction was carried out at 37 °C for 1 h and quenched by the addition of acidic PCI, followed by RNA purification. Finally, the integrity of the capped mRNAs was verified by agarose gel electrophoresis.
Preparation of HeLa translation-competent lysates
HeLa S3 lysates were prepared similarly to the description in ref. 14. Briefly, lysates were prepared from S3 HeLa cell cultures grown to a cell density ranging from 1 × 106 to 2 × 106 cells per ml. Cells were pelleted (200g, 4 °C for 5 min) and washed twice with cold PBS pH 7.4 and finally resuspended in ice-cold hypotonic lysis buffer (10 mM HEPES pH 7.3, 10 mM K-acetate, 500 μM Mg-acetate, 5 mM DTT and 1× protease inhibitor cocktail (biotool.com)) at a final concentration of 2 × 108 cells per ml. The suspension was incubated on ice for 10 min and cells were lysed by dual centrifugation (500 r.p.m., −5 °C, 4 min) using Zentrimix 380 R system (Hettich) with a 3206 rotor and 3209 adapters. The lysis process was monitored by trypan staining. The lysate was centrifuged at 13,000g, 4 °C for 10 min and the supernatant was aliquoted, snap-frozen and stored at −80 °C.
In vitro translation assays
In vitro translation reactions were performed similarly to the description in ref. 14. Briefly, 400 μl of recombinant proteins were dialyzed overnight in 30 mM NaCl, 5 mM HEPES pH 7.3 at 4 °C using Slide-A-Lyzer MINI Dialysis devices with a 3.5k MWCO (Thermo Scientific, 88400), and the protein concentration was calculated using Nanodrop. In parallel, equal volumes of recombinant protein storage buffer were dialyzed and used as negative control (0 μM condition) and to maintain the same concentration of dialyzed storage buffer in translation mixtures. S3 lysate corresponding to 1.11 × 106 cell equivalents was used at a concentration of 8.88 × 107 cell equivalents per ml. The reaction was supplemented to a final concentration of 15 mM HEPES, pH 7.3, 0.3 mM MgCl2, 24 mM KCl, 28 mM K-acetate, 6 mM creatine phosphate (Roche), 102 ng µl−1 creatine kinase (Roche), 0.4 mM amino acid mixture (Promega) and 1 U µl−1 NxGen RNase inhibitor (Lucigen). Control reactions contained 320 µg ml−1 puromycin (Santa Cruz Biotechnology) and all reactions were complemented with an equal volume of dialyzed protein purification buffer. In all reactions where recombinant Nsp1 was used, the lysate was pre-incubated with Nsp1 at 4 °C for 30 min. Before addition of reporter mRNAs, the mixtures were incubated at 33 °C for 5 min. In vitro transcribed and capped mRNAs were incubated for 5 min at 65 °C, 15 min at room temperature (RT) and cooled on ice. Reporter mRNAs were added to the translation reactions at a final concentration of 40 fmol μl−1. The translation reaction was performed at 33 °C for 50 min. To monitor the protein synthesis output, samples were put on ice and mixed with 50 μl 1× Renilla-Glo substrate (Promega) in Renilla-Glo (Promega) assay buffer on a white-bottomed 96-well plate. The plate was incubated at 30 °C for 10 min and the luminescence signal was measured three times using the TECAN infinite M100 Pro plate reader and plotted on GraphPad after performing three independent biological replicates.
Transient expression of FLAG-Nsp1 in HeLa cells and puromycin incorporation assay
The plasmid encoding the N-terminally tagged WT Nsp1 DNA sequence (pcDNA3.1(+) backbone) was ordered from GeneScript and the mutant K164A/H165A (KH) construct was created by site-directed mutagenesis using the primer 5′-GTT CCC TTG TCA CGC CGC TAC TAG CTG CGG TAT TCC AAT TCT CCT GAA AAT-3′.
HeLa cells were cultured in DMEM supplemented with 10% FCS supplemented with 100 UI ml−1 penicillin and 100 μg ml−1 streptomycin (DMEM+/+) at 37 °C, 5% CO2. To compare expression levels of different FLAG-Nsp1 protein variants (WT/KH), ~4 × 105 cells were transfected with 400 ng of each plasmid using 7.5 μl μg−1 DNA Dogtor transfection reagent (Oz Biosciences) in Opti-MEM (Thermo Fisher Scientific). As a control, an equal amount of empty vector was added to the reaction. After 24 h the samples were collected by trypsinization, counted and lysed in RIPA buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 1% SDS). The whole-cell extract was then isolated by centrifugation at 13,000g, 4 °C. Finally, the extracts were combined with an equal volume of 2× LDS sample buffer (Invitrogen) containing 100 mM DTT.
For the puromycin incorporation assay, ~1.25 × 105 cells were transfected with 500 ng of the WT FLAG-Nsp1 plasmid (or empty vector) and 100 ng of the FLAG-Nsp1 KH mutant plasmid (using Dogtor and Opti-MEM as above). After one day, the transfection was repeated with double the amount of plasmid DNA and the cells were incubated for 24 h again. Forty minutes before the end of this incubation time the medium was changed to DMEM+/+ containing 10 μg ml−1 puromycin (Santa Cruz Biotechnology) for 10 min to label newly synthesized proteins. Subsequently, the cells were washed with 1× PBS and the cells were recovered in the original medium for 30 min. Ultimately, the cells were collected as described above. In addition, a positive control condition for translation inhibition was included (empty vector transfected) in which the medium was supplemented with CHX (Focus Biomolecules) to 100 μg ml−1 within the last 2 h of the experiment (apart from the puromycin pulse).
From the prepared protein samples, 2 × 105 cell equivalents were resolved by SDS-PAGE (NuPAGE 4 to 12%, Bis-Tris, 1.0 mm, Midi Protein Gel, 12 + 2-well, Thermo Fisher) in MOPS buffer. For analysis of incorporated puromycin, an additional 1:2 dilution was loaded. Proteins were then blotted on a nitrocellulose membrane (iBlot 2, NC Regular Stacks, program P0), blocked and then probed with the following primary antibodies: mouse anti-puromycin (EMD Milllipore, cat. no. MABE343, used at 1:15,000), mouse anti-FLAG M2 (Sigma Aldrich, cat. no. F1804, used at 1:2,500) and rabbit anti-GAPDH (Santa Cruz Biotechnology, cat. no. sc-25778, used at 1:10,000). The signal on the anti-puromycin-stained blot was quantified using ImageJ 1.52p with the line selection tool to measure the whole lane intensity of the puromycin signal (manual background subtraction) and the gel analyzer tool to measure the intensity of the GAPDH loading control.
HEK extracts and sucrose gradient analysis
HEK293E cell lysates were supplemented with Nsp1 to purify native-like Nsp1-inhibited translation complexes. For this, frozen HEK293E cells were thawed and resuspended in 2× excess of lysis buffer (25 mM HEPES-KOH pH 7.6, 5 mM MgCl2, 50 mM KCl, cOmplete protease inhibitor cocktail (Roche), 1 mM PMSF, 0.2 U µl−1 RiboLock). For cell lysis, cells were transferred to a Dounce homogenizer (tight) and lysed with 12 strokes. After adding Triton X-100 to a final concentration of 0.1%, the lysate was incubated under rotation for 30 min at 4 °C and cleared for 10 min in an MLA-80 rotor (Beckman Coulter) at 11,500g and 4 °C. The cleared HEK293E lysate was supplemented with untagged Nsp1 (for cryo-EM) or His6-Nsp1 (for western blot) to a final concentration of 2 µM, and the extracts were incubated for an additional 5 min at 30 °C before they were loaded onto 15–45% (wt/vol) sucrose gradients (20 mM HEPES-KOH pH 7.6, 100 mM KOAc, 5 mM MgCl2, 1 mM DTT). Gradients were centrifuged in a SW 32.1 Ti rotor (Beckman) at 79,500g for 15 h at 4 °C and manually fractioned with a syringe. Fractions containing ribosomal particles were pooled and concentrated in an Amicon Ultra-15 centrifugal filter (100-kDa MWCO). For biochemical analyses, the same gradients were prepared, with the exception of using His6-tagged Nsp1 instead of the TEV-cleaved protein. Fractions were precipitated with trichloroacetic acid (TCA) and subjected to western blot analysis (anti-His antibody, Clontech). Additionally, before precipitation, samples were taken for analysis on agarose gels (0.06% bleach, 1% (wt/vol) agarose).
Cryo-electron microscopy sample preparation and data collection
Quantifoil R2/2 holey carbon copper grids (Quantifoil Micro Tool) were prepared by first applying an additional thin layer of continuous carbon and then glow-discharging them for 15 s at 15 mA using an easiGlow Discharge cleaning system (PELCO). For the in vitro binding experiment, purified Nsp1 was first mixed with 40S in a molar ratio of 10:1. For the HEK lysate sample, sucrose peak fractions containing ribosomes were collected, buffer exchanged and concentrated, then, 4-µl samples at concentrations of 80–100 nM of the 40S–Nsp1 or ribosomes from the HEK cell lysate were applied to the grids, which were then blotted for ~8 s and immediately plunged in 1:2 ethane:propane (Carbagas) at liquid nitrogen temperature using a Vitrobot sytem (Thermo Fisher Scientific). The Vitrobot chamber was kept at 4 °C and 100% humidity during the whole procedure.
For each sample, one grid was selected for data collection using a Titan Krios cryo-transmission electron microscope (Thermo Fisher Scientific) operating at 300 kV and equipped with a K3 camera (Gatan), which was run in counting and super-resolution mode, mounted to a GIF Quantum LS imaging filter operated with an energy filter slit width of 20 eV. The K3 datasets were collected at a nominal magnification of ×81,000 (physical pixel size of 1.08 Å per pixel, which corresponds to a super-resolution pixel size of 0.54 Å per pixel). For the counting mode, illumination conditions were adjusted to an exposure rate of 24 e− pixel−1 s−1. Micrographs were recorded as video stacks at an electron dose of ~60 e− Å−2 applied over 40 frames. For both datasets, the defocus was varied from ~−1 to −3 μm.
Cryo-electron microscopy data processing
The stacks of frames were first aligned to correct for motion during exposure, dose-weighted and gain-corrected using MotionCor2 (ref. 26). The super-resolution micrographs collected with the K3 camera were additionally binned twice during the MotionCor2 procedure. The contrast transfer functions of the motion-corrected and dose-weighted micrographs were then estimated using GCTF27.
Micrographs (10,104 for the in vitro binding experiment, 15,866 for the HEK cell extract) were carefully inspected based on CTF estimations for drift and ice quality. Particle images of ribosomes were picked (2,078,577 for the in vitro binding experiment, 1,358,638 for the HEK cell extract) in RELION3.1 using a Laplacian-of-Gaussian filter-based method28. The picked particle images were then subjected to a reference-free two-dimensional (2D) classification in RELION/cryoSPARC2, and the particles were selected from the 2D class averages (1,718,196 particles for the in vitro binding experiment, 1,113,915 for the HEK cell extract). For the in vitro binding experiment, the particles were then classified in 3D using a human 40S reinitiation complex (EMD 3770 (ref. 25)) that was low-pass-filtered to 60 Å to select for good 40S classes, followed by refinement using RELION3.1 (ref. 29). Further processing was done with RELION3.1 and cryoSPARC2 (ref. 30) according to the scheme shown in Extended Data Fig. 2. Transformation of particle information between the two programs was done using PyEM script (https://doi.org/10.5281/zenodo.3576630). In short, the particle set was first cleaned from the preferentially oriented particles based on their orientation parameters, which reduced the particle set to 700,459 particles. Those particles were then further classified for their quality and for the presence of Nsp1 using a focused 3D classification approach. The final set of particle images was refined using a global 3D refinement. To further improve the local resolution of the 40S–Nsp1 complex, masks around the 40S head and body were generated using UCSF Chimera31 by creating a mask that was extended by 10 Å around a fitted model of the 40S subunit. Those masks were used for a multi-body refinement in RELION3.1 (ref. 32). Finally, the two focused maps were combined to generate a composite 3D map of the entire in vitro reconstituted 40S–Nsp1 complex.
For the HEK cell extract, after 2D classifications, ab initio reconstruction was performed in cryoSPARC2 (ref. 30), and the determined volumes were used as starting references for a heterogeneous refinement in cryoSPARC2 (Extended Data Fig. 1). The 204,114 particle images corresponding to the 40S ribosomal subunit were selected for a further round of heterogeneous refinement in cryoSPARC2, which resolved a density corresponding to initiation factor eIF3 in a fraction of the particles. To improve the occupancy of eIF3, particle images belonging to the 40S subunit class were then subjected to a focused 3D classification in RELION3.1 using a circular mask on the eIF3 region. The 3D class depicting the best density for eIF3 was selected (18,692 particle images) and was then used for a global 3D refinement.
Structure building and refinement
For building of the 40S–Nsp1 complex, the head and body of PDB 5OA3 (ref. 25) were docked as rigid bodies into the 2.8-Å head and body maps that were obtained by focused classification (Extended Data Fig. 2). The structures were adjusted manually into the high-resolution maps using COOT33, and the C terminus of Nsp1 (residues 148–180), which was well-resolved in the map of the 40S body, was built de novo. The coordinates were subjected to five cycles of real-space refinement using PHENIX 1.18 (ref. 34). To stabilize the refinement in less well-resolved peripheral areas, protein secondary structure and Ramachandran as well as RNA base pair restraints were applied. Remaining discrepancies between models and maps as well as missing Mg2+ ions were detected using real-space difference maps and, after model completion, the coordinates were refined for two additional cycles. The resulting final models have excellent geometries and correlations between the maps and models (Table 1 and Extended Data Fig. 2). The structures were validated using MolProbity35 and by comparison of the model versus map FSCs at values of 0.5, which coincided well with the FSCs between the half-sets of the EM reconstruction using the FSC = 1.43 criterion (Extended Data Fig. 2).
To assemble the full 40S–Nsp1 complex, both refined structures were docked into a 2.8-Å chimeric map comprising the complete 40S–Nsp1. After readjustment of the head-to-body connections, the complete model was subjected to two additional rounds of real-space refinement as described above.
The 4.1-Å and 2.5-Å maps of the Nsp1–43S PIC and non-translating 80S shown in Extended Data Figs. 1 and 3 were aligned onto the 2.8-Å 40S–Nsp1 body map of the in vitro reconstituted complex in UCSF Chimera, into which the atomic model had been built. For general interpretation of 43S PIC and non-translating 80S, the refined models of the 40S head and body determined for the in vitro 40S–Nsp1 complex were docked as rigid bodies in UCSF Chimera31. To highlight the regions of the initiation factors in the cryo-EM map, initiation factors IF2 and IF3 were taken from PDB 6YAM36 and docked similarly. A homology model for the missing eIF2β subunit was obtained using PHYRE2 (ref. 37) and PDB 3JAP38 as a template, and the density of IF1 was interpreted using PDB 2IF1 (ref. 39).
Reporting Summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this Article.
Data availability
Cryo-EM maps and atomic models have been deposited in the Electron Microscopy Data Bank (EMDB) and wwPDB, respectively, with the following accession codes: EMD-11320 and PDB 6ZOJ (SARS-CoV-2–Nsp1–40S, composite map); EMD-11321 and PDB 6ZOK (SARS-CoV-2–Nsp1–40S complex, focused on body); EMD-11322 and PDB 6ZOL (SARS-CoV-2–Nsp1–40S complex, focused on head). In addition, the maps for 43S PIC–Nsp1 and non-translating 80S–Nsp1 have been deposited in the EMDB as EMD-11323 and EMD-11609, respectively. Source data are provided with this paper.
Change history
20 October 2020
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
Gorbalenya, A. E. et al. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 5, 536–544 (2020).
Lim, Y., Ng, Y., Tam, J. & Liu, D. Human coronaviruses: a review of virus–host interactions. Diseases 4, 26 (2016).
Prentice, E., McAuliffe, J., Lu, X., Subbarao, K. & Denison, M. R. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J. Virol. 78, 9977–9986 (2004).
Nakagawa, K., Lokugamage, K. G. & Makino, S. Viral and cellular mRNA translation in coronavirus-infected cells. Adv. Virus Res 96, 165–192 (2016).
Thiel, V. et al. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 84, 2305–2315 (2003).
Kamitani, W. et al. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc. Natl Acad. Sci. USA 103, 12885–12890 (2006).
Kamitani, W., Huang, C., Narayanan, K., Lokugamage, K. G. & Makino, S. A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat. Struct. Mol. Biol. 16, 1134–1140 (2009).
Huang, C. et al. SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: viral mRNAs are resistant to nsp1-induced RNA cleavage. PLoS Pathog. 7, e1002433 (2011).
Lokugamage, K. G., Narayanan, K., Huang, C. & Makino, S. Severe acute respiratory syndrome coronavirus protein nsp1 is a novel eukaryotic translation inhibitor that represses multiple steps of translation initiation. J. Virol. 86, 13598–13608 (2012).
Almeida, M. S., Johnson, M. A., Herrmann, T., Geralt, M. & Wüthrich, K. Novel β-barrel fold in the nuclear magnetic resonance structure of the replicase nonstructural protein 1 from the severe acute respiratory syndrome coronavirus. J. Virol. 81, 3151–3161 (2007).
Narayanan, K. et al. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J. Virol. 82, 4471–4479 (2008).
Wathelet, M. G., Orr, M., Frieman, M. B. & Baric, R. S. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J. Virol. 81, 11620–11633 (2007).
Züst, R. et al. Coronavirus non-structural protein 1 is a major pathogenicity factor: implications for the rational design of coronavirus vaccines. PLoS Pathog. 3, 1062–1072 (2007).
Karousis, E. D., Gurzeler, L.-A., Annibaldis, G., Dreos, R. & Mühlemann, O. Human NMD ensues independently of stable ribosome stalling. Nat. Commun. https://doi.org/10.1038/s41467-020-17974-z (2020).
Ceraolo, C. & Giorgi, F. M. Genomic variance of the 2019-nCoV coronavirus. J. Med. Virol. 92, 522–528 (2020).
Lokugamage, K. G. et al. Middle East respiratory syndrome coronavirus nsp1 inhibits host gene expression by selectively targeting mRNAs transcribed in the nucleus while sparing mRNAs of cytoplasmic origin. J. Virol. 89, 10970–10981 (2015).
Brown, A., Baird, M. R., Yip, M. C. J., Murray, J. & Shao, S. Structures of translationally inactive mammalian ribosomes. Elife 7, e40486 (2018).
Anger, A. M. et al. Structures of the human and Drosophila 80S ribosome. Nature 497, 80–85 (2013).
Yang, D. & Leibowitz, J. L. The structure and functions of coronavirus genomic 3′ and 5′ ends. Virus Res. 206, 120–133 (2015).
van den Born, E., Posthuma, C. C., Gultyaev, A. P. & Snijder, E. J. Discontinuous subgenomic RNA synthesis in arteriviruses is guided by an RNA hairpin structure located in the genomic leader region. J. Virol. 79, 6312–6324 (2005).
Rangan, R., Zheludev, I. N. & Das, R. RNA genome conservation and secondary structure in SARS-CoV-2 and SARS-related viruses: a first look. RNA 26, 937–959 (2020); https://doi.org/10.1261/rna.076141.120
Kim, D. et al. The architecture of SARS-CoV-2 transcriptome. Cell 181, 914–921 (2020).
Thoms, M. et al. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science https://doi.org/10.1126/science.abc8665 (2020).
de Lima Menezes, G. & da Silva, R. A. Identification of potential drugs against SARS-CoV-2 non-structural protein 1 (nsp1). J. Biomol. Struct. Dyn. 2020, 1–11 (2020).
Weisser, M. et al. Structural and functional insights into human re-initiation complexes. Mol. Cell 67, 447–456 (2017).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Egelman, E. H. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife https://doi.org/10.7554/eLife.42166.001 (2018).
Scheres, S. H. W. & Chen, S. Prevention of overfitting in cryo-EM structure determination. Nat. Methods 9, 853–854 (2012).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. CryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Meng, E. C., Pettersen, E. F., Couch, G. S., Huang, C. C. & Ferrin, T. E. Tools for integrated sequence-structure analysis with UCSF Chimera. BMC Bioinformatics 7, 339 (2006).
Nakane, T., Kimanius, D., Lindahl, E. & Scheres, S. H. Characterisation of molecular motions in cryo-EM single-particle data by multi-body refinement in RELION Elife https://doi.org/10.7554/eLife.36861 (2018).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75, 861–877 (2019).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Simonetti, A., Guca, E., Bochler, A., Kuhn, L. & Hashem, Y. Structural insights into the mammalian late-stage initiation complexes. Cell Rep. 31, 107497 (2020).
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N. & Sternberg, M. J. E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015).
Llácer, J. L. et al. Conformational differences between open and closed states of the eukaryotic translation initiation complex. Mol. Cell 59, 399–412 (2015).
Fletcher, C. M., Pestova, T. V., Hellen, C. U. T. & Wagner, G. Structure and interactions of the translation initiation factor eIF1. EMBO J. 18, 2631–2637 (1999).
Huston, N. C. et al. Comprehensive in-vivo secondary structure of the SARS-CoV-2 genome reveals novel regulatory motifs and mechanisms. Preprint at bioRxiv https://www.biorxiv.org/content/10.1101/2020.07.10.197079v1 (2020).
Acknowledgements
We thank the ETH Scientific Center for optical and electron microscopy (ScopeM) and the CryoEM Knowledge (CEMK) hub, in particular D. Böhringer, for technical support and the opportunity to continue our work in spite of the ETH lockdown due to the COVID-19 pandemic. We thank the Functional Genomics Center Zurich (FGCZ) for help with MS. We thank their teams for the support in the lab, and especially M. Jia, P. Bhatt and D. Yudin for creating a productive working atmosphere. This work was supported by grants to N.B., O.M. and V.T. from the Swiss National Science Foundation (SNSF; grants 173085, 182341 and 182831); the National Center of Competence in Research (NCCR) on RNA and Disease funded by the SNSF; ETH research grant ETH-23 18-2; and a PhD fellowship from Böhringer Ingelheim Fonds to K.S.
Author information
Authors and Affiliations
Contributions
N.B. and K.S. initiated the project and designed the experiments. K.S. expressed proteins, together with B.E., and prepared samples for cryo-EM. K.S., A.J. and A.S. prepared grids and carried out data collection and processing. E.D.K. and O.M. designed translation experiments. E.D.K. and L.-A.G. were involved in cloning, and E.D.K. performed in vitro translation reactions, with the help of L.-A.G. L.-A.G. performed the puromycin incorporation assay. K.S. and B.E. performed sucrose binding assays. M.L. was involved in structure modeling and refinement as well as in figure preparation. N.B. and K.S. coordinated the project. V.T. contributed to data interpretation and all authors contributed to the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Anke Sparmann was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Data processing of the HEK cell extract cryo-EM dataset.
Scheme for the processing of the HEK cell extract sample. Local resolution estimates are plotted as heat map on the final volumes accompanied with a slice through the volume. The half map vs. half map FSC curves are shown for the refinement of the 43S PIC (cyan) and the refinement of the non-translating 80S complex (purple).
Extended Data Fig. 2 Data processing of the in vitro reconstituted 40S-Nsp1 cryo-EM dataset.
Scheme of the processing steps performed for the sample of the in vitro binding experiment. The local resolution estimate is plotted on the final volumes as heat map, together with additional slices through the volumes. The half map vs. half map FSC curves are plotted for the overall refinement (purple), the refinement focused on the body (blue), on the head (cyan), as well as for the composite map (green). The map vs. model FSCs are plotted for the body (yellow) and the head (orange) in their respective focused maps, as well as for the full 40S in the composite map (red).
Extended Data Fig. 3 Binding of Nsp1 to the 43S pre-initiation and 80S complexes.
Additional EM density is present in the 40S mRNA entry site. Docking of the high-resolution 40S-Nsp1 structure into the 4.1 Å map of the 43S PIC (a) and into the 2.5 Å map of the 80S complex (c) reveal excellent fits of the two C-terminal Nsp1 helices. For several bulky side chains mediating the hydrophobic contact with uS5 of the 40S body, side chain densities can be recognized in the 43S PIC (b) and 80S complexes (d).
Extended Data Fig. 4 Sucrose pelleting binding assay of Nsp1 mutants with the C-terminal domain removed or the N-terminal domain exchanged with the soluble domain of E.coli thioredoxin.
In the in vitro binding assay, the N-term – linker only Nsp1 (a) and the Trx1 – linker – C-term Nsp1 (b) mutants were added to 40S and 60S ribosomal SU and loaded on a 30% (wt/vol) sucrose cushion. Unbound proteins remained in the supernatant (SN), while bound proteins co-pelleted with 40S (P). (c) The same Western blot gel for WT Nsp1 as in Fig. 1d is depicted for comparison. Uncropped gel images for a-c are available online as Source data.
Extended Data Fig. 5 Components of in vitro translation reaction.
(a) Predicted secondary structure of the 5’ region of SARS-CoV-2 genomic RNA (based on21,40) with 5’ UTR depicted in blue, the protein encoding sequence in black and the transcriptional regulatory sequence (TRS) in orange. Stem loops (SL) are labeled. (b) 1% agarose gel electrophoresis of the in vitro transcribed RLuc and FL-RLuc reporters used for the in vitro translation assays. (c) Input samples of the in vitro translation assay described in Fig. 3b. 2 μL of dialyzed Nsp1 protein samples at a concentration of 17.9 μM were analyzed on a 4–12% SDS-PAGE and stained by Imperial protein staining (Thermo Fisher Scientific).
Supplementary information
Supplementary Information
Supplementary Fig. 1.
Source data
Source Data Fig. 1
Unprocessed western blots and gels.
Source Data Fig. 3
Unprocessed western blots.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Unprocessed western blots.
Source Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 4
Unprocessed western blots.
Rights and permissions
About this article
Cite this article
Schubert, K., Karousis, E.D., Jomaa, A. et al. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat Struct Mol Biol 27, 959–966 (2020). https://doi.org/10.1038/s41594-020-0511-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-020-0511-8