Abstract
During liver regeneration, most new hepatocytes arise via self-duplication; yet, the underlying mechanisms that drive hepatocyte proliferation following injury remain poorly defined. By combining high-resolution transcriptome and polysome profiling of hepatocytes purified from quiescent and toxin-injured mouse livers, we uncover pervasive alterations in messenger RNA translation of metabolic and RNA-processing factors, which modulate the protein levels of a set of splicing regulators. Specifically, downregulation of the splicing regulator ESRP2 activates a neonatal alternative splicing program that rewires the Hippo signaling pathway in regenerating hepatocytes. We show that production of neonatal splice isoforms attenuates Hippo signaling, enables greater transcriptional activation of downstream target genes, and facilitates liver regeneration. We further demonstrate that ESRP2 deletion in mice causes excessive hepatocyte proliferation upon injury, whereas forced expression of ESRP2 inhibits proliferation by suppressing the expression of neonatal Hippo pathway isoforms. Thus, our findings reveal an alternative splicing axis that supports regeneration following chronic liver injury.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
All raw RNA-seq and polysome profiling data files are available for download from the Gene Expression Omnibus under accessions GSE105944 and GSE106140. Data underlying the analyses in Figs. 1f–h, 2d, 3c,e, and 4a, and Supplementary Fig. 3j are available in Supplementary Dataset 2. Source data for Figs. 3a, 5d–h, and 7d, e, and Supplementary Figs. 4 and 7 are available in Supplementary Dataset 3. All other data are available from the authors upon reasonable request.
References
Michalopoulos, G. K. & DeFrances, M. C. Liver regeneration. Science 276, 60–66 (1997).
Taub, R. Liver regeneration: from myth to mechanism. Nat. Rev. Mol. Cell Biol. 5, 836–847 (2004).
Michalopoulos, G. K. Liver regeneration. J. Cell. Physiol. 213, 286–300 (2007).
Forbes, S. J. & Newsome, P. N. Liver regeneration—Mechanisms and models to clinical application. Nat. Rev. Gastroenterol. Hepatol. 13, 473–485 (2016).
Miyaoka, Y. et al. Hypertrophy and unconventional cell division of hepatocytes underlie liver regeneration. Curr. Biol. 22, 1166–1175 (2012).
Lu, W. Y. et al. Hepatic progenitor cells of biliary origin with liver repopulation capacity. Nat. Cell Biol. 17, 971–983 (2015).
Raven, A. et al. Cholangiocytes act as facultative liver stem cells during impaired hepatocyte regeneration. Nature 547, 350–354 (2017).
Miyajima, A., Tanaka, M. & Itoh, T. Stem/progenitor cells in liver development, homeostasis, regeneration, and reprogramming. Cell Stem Cell 14, 561–574 (2014).
Michalopoulos, G. K. & Khan, Z. Liver stem cells: experimental findings and implications for human liver disease. Gastroenterology 149, 876–882 (2015).
Espanol-Suner, R. et al. Liver progenitor cells yield functional hepatocytes in response to chronic liver injury in mice. Gastroenterology 143, 1564–1575.e7 (2012).
Malato, Y. et al. Fate tracing of mature hepatocytes in mouse liver homeostasis and regeneration. J. Clin. Invest. 121, 4850–4860 (2011).
Schaub, J. R., Malato, Y., Gormond, C. & Willenbring, H. Evidence against a stem cell origin of new hepatocytes in a common mouse model of chronic liver injury. Cell Rep. 8, 933–939 (2014).
Tarlow, B. D. et al. Bipotential adult liver progenitors are derived from chronically injured mature hepatocytes. Cell Stem Cell 15, 605–618 (2014).
Yanger, K. et al. Adult hepatocytes are generated by self-duplication rather than stem cell differentiation. Cell Stem Cell 15, 340–349 (2014).
Jors, S. et al. Lineage fate of ductular reactions in liver injury and carcinogenesis. J. Clin. Invest. 125, 2445–2457 (2015).
Font-Burgada, J. et al. Hybrid periportal hepatocytes regenerate the injured liver without giving rise to cancer. Cell 162, 766–779 (2015).
Preisegger, K. H. et al. Atypical ductular proliferation and its inhibition by transforming growth factor beta1 in the 3,5-diethoxycarbonyl-1,4-dihydrocollidine mouse model for chronic alcoholic liver disease. Lab. Invest. 79, 103–109 (1999).
Fickert, P. et al. A new xenobiotic-induced mouse model of sclerosing cholangitis and biliary fibrosis. Am. J. Pathol. 171, 525–536 (2007).
Bhate, A. et al. ESRP2 controls an adult splicing programme in hepatocytes to support postnatal liver maturation. Nat. Commun. 6, 8768 (2015).
Young, S. K., Willy, J. A., Wu, C., Sachs, M. S. & Wek, R. C. Ribosome reinitiation directs gene-specific translation and regulates the integrated stress response. J. Biol. Chem. 290, 28257–28271 (2015).
Maslon, M. M., Heras, S. R., Bellora, N., Eyras, E. & Caceres, J. F. The translational landscape of the splicing factor SRSF1 and its role in mitosis. Elife, e02028 (2014).
Baralle, F. E. & Giudice, J. Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 18, 437–451 (2017).
Braunschweig, U., Gueroussov, S., Plocik, A. M., Graveley, B. R. & Blencowe, B. J. Dynamic integration of splicing within gene regulatory pathways. Cell 152, 1252–1269 (2013).
Fu, X. D. & Ares, M. Jr. Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet. 15, 689–701 (2014).
Kalsotra, A. & Cooper, T. A. Functional consequences of developmentally regulated alternative splicing. Nat. Rev. Genet. 12, 715–729 (2011).
Lee, Y. & Rio, D. C. Mechanisms and regulation of alternative pre-mRNA splicing. Annu. Rev. Biochem. 84, 291–323 (2015).
Papasaikas, P. & Valcarcel, J. The spliceosome: the ultimate RNA chaperone and sculptor. Trends. Biochem. Sci. 41, 33–45 (2016).
Yang, X. et al. Widespread expansion of protein interaction capabilities by alternative splicing. Cell 164, 805–817 (2016).
Buljan, M. et al. Tissue-specific splicing of disordered segments that embed binding motifs rewires protein interaction networks. Mol. Cell 46, 871–883 (2012).
Ellis, J. D. et al. Tissue-specific alternative splicing remodels protein–protein interaction networks. Mol. Cell 46, 884–892 (2012).
Irimia, M. et al. A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell 159, 1511–1523 (2014).
Ray, D. et al. Rapid and systematic analysis of the RNA recognition specificities of RNA-binding proteins. Nat. Biotechnol. 27, 667–670 (2009).
Bebee, T. W. et al. The splicing regulators Esrp1 and Esrp2 direct an epithelial splicing program essential for mammalian development. eL ife 4, https://doi.org/10.7554/eLife.08954 (2015).
Dittmar, K. A. et al. Genome-wide determination of a broad ESRP-regulated posttranscriptional network by high-throughput sequencing. Mol. Cell. Biol. 32, 1468–1482 (2012).
Yu, F. X., Zhao, B. & Guan, K. L. Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell 163, 811–828 (2015).
Johnson, R. & Halder, G. The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat. Rev. Drug Discov. 13, 63–79 (2014).
Harvey, K. F., Zhang, X. & Thomas, D. M. The Hippo pathway and human cancer. Nat. Rev. Cancer 13, 246–257 (2013).
Benhamouche, S. et al. Nf2/Merlin controls progenitor homeostasis and tumorigenesis in the liver. Genes Dev. 24, 1718–1730 (2010).
Lu, L. et al. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc. Natl Acad. Sci. USA 107, 1437–1442 (2010).
Yimlamai, D. et al. Hippo pathway activity influences liver cell fate. Cell 157, 1324–1338 (2014).
Zhang, N. et al. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev. Cell 19, 27–38 (2010).
Bai, H. et al. Yes-associated protein regulates the hepatic response after bile duct ligation. Hepatology 56, 1097–1107 (2012).
Grijalva, J. L. et al. Dynamic alterations in Hippo signaling pathway and YAP activation during liver regeneration. Am. J. Physiol. Gastrointest. Liver Physiol. 307, G196–G204 (2014).
Su, T. et al. Two-signal requirement for growth-promoting function of Yap in hepatocytes. eL ife 4, https://doi.org/10.7554/eLife.02948 (2015).
Sher, I., Hanemann, C. O., Karplus, P. A. & Bretscher, A. The tumor suppressor merlin controls growth in its open state, and phosphorylation converts it to a less-active more-closed state. Dev. Cell 22, 703–705 (2012).
Li, Y. et al. Angiomotin binding-induced activation of Merlin/NF2 in the Hippo pathway. Cell Res. 25, 801–817 (2015).
Graves, P. R. & Roach, P. J. Role of COOH-terminal phosphorylation in the regulation of casein kinase I delta. J. Biol. Chem. 270, 21689–21694 (1995).
Rivers, A., Gietzen, K. F., Vielhaber, E. & Virshup, D. M. Regulation of casein kinase I epsilon and casein kinase I delta by an in vivo futile phosphorylation cycle. J. Biol. Chem. 273, 15980–15984 (1998).
Cegielska, A., Gietzen, K. F., Rivers, A. & Virshup, D. M. Autoinhibition of casein kinase I epsilon (CKI epsilon) is relieved by protein phosphatases and limited proteolysis. J. Biol. Chem. 273, 1357–1364 (1998).
Gaffney, C. J. et al. Identification, basic characterization and evolutionary analysis of differentially spliced mRNA isoforms of human YAP1 gene. Gene 509, 215–222 (2012).
Sudol, M. YAP1 oncogene and its eight isoforms. Oncogene 32, 3922 (2013).
Finch-Edmondson, M. L., Strauss, R. P., Clayton, J. S., Yeoh, G. C. & Callus, B. A. Splice variant insertions in the C-terminus impairs YAP’s transactivation domain. Biochem. Biophys. Rep. 6, 24–31 (2016).
Lin, K. C., Park, H. W. & Guan, K. L. Regulation of the Hippo pathway transcription factor TEAD. Trends Biochem. Sci. 42, 862–872 (2017).
Anbanandam, A. et al. Insights into transcription enhancer factor 1 (TEF-1) activity from the solution structure of the TEA domain. Proc. Natl Acad. Sci. USA 103, 17225–17230 (2006).
Jiang, S. W., Dong, M., Trujillo, M. A., Miller, L. J. & Eberhardt, N. L. DNA binding of TEA/ATTS domain factors is regulated by protein kinase C phosphorylation in human choriocarcinoma cells. J. Biol. Chem. 276, 23464–23470 (2001).
Gupta, M. P., Kogut, P. & Gupta, M. Protein kinase-A dependent phosphorylation of transcription enhancer factor-1 represses its DNA-binding activity but enhances its gene activation ability. Nucleic Acids Res. 28, 3168–3177 (2000).
Wu, J. C., Merlino, G. & Fausto, N. Establishment and characterization of differentiated, nontransformed hepatocyte cell lines derived from mice transgenic for transforming growth factor alpha. Proc. Natl Acad. Sci. USA 91, 674–678 (1994).
Meng, Z., Moroishi, T. & Guan, K. L. Mechanisms of Hippo pathway regulation. Genes Dev. 30, 1–17 (2016).
Dupont, S. et al. Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183 (2011).
Dong, J. et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130, 1120–1133 (2007).
Li, W. C., Ralphs, K. L. & Tosh, D. Isolation and culture of adult mouse hepatocytes. Methods Mol. Biol. 633, 185–196 (2010).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome. Biol. 15, 550 (2014).
Shen, S. H. et al. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl Acad. Sci. USA 111, E5593–E5601 (2014).
Park, J. W., Jung, S., Rouchka, E. C., Tseng, Y. T. & Xing, Y. rMAPS: RNA map analysis and plotting server for alternative exon regulation. Nucleic Acids Res. 44, W333–W338 (2016).
Huang da, W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Tranchevent, L. C. et al. Identification of protein features encoded by alternative exons using Exon Ontology. Genome Res. 27, 1087–1097 (2017).
Schneider-Poetsch, T. et al. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat. Chem. Biol. 6, 209–217 (2010).
Chorghade, S. et al. Poly(A) tail length regulates PABPC1 expression to tune translation in the heart. eLife 6, https://doi.org/10.7554/eLife.24139 (2017).
Floor, S. N. & Doudna, J. A. Tunable protein synthesis by transcript isoforms in human cells. eLife 5, https://doi.org/10.7554/eLife.10921 (2016).
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012).
Wagner, G. P., Kin, K. & Lynch, V. J. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 131, 281–285 (2012).
Kalsotra, A., Wang, K., Li, P. F. & Cooper, T. A. MicroRNAs coordinate an alternative splicing network during mouse postnatal heart development. Genes Dev. 24, 653–658 (2010).
Acknowledgements
We thank the members of the Kalsotra and Anakk laboratories for their valuable discussions and comments on the manuscript. This research was supported through NIH (R01HL126845) and March of Dimes (5-FY14-112) grants to A.K. and NIH (R01AI081710) to S.A. W.A. was supported by the NIH predoctoral NRSA fellowship (F30DK108567). J.S. was partly supported by the NIH Chemistry–Biology Interface Training Grant (5T32-GM070421) and the American Heart Association predoctoral fellowship (17PRE33670030). A.B. was partly supported by the Herbert E. Carter fellowship in Biochemistry, UIUC. Three cores at UIUC supported this project: Transgenic Mouse Facility Core, High-Throughput Sequencing and Genotyping Core, and Histology and Microscopy Core.
Author information
Authors and Affiliations
Contributions
S.B., W.A., J.S., and A.K. conceived the project and designed the experiments. S.B., W.A., J.S., A.B., J.C., and E.H.R. performed experiments. R.P.C. and S.A. provided reagents. S.B., W.A., J.S., and A.K. interpreted results and wrote the manuscript. All authors discussed the results and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
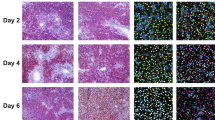
Supplementary Figure 1 Validation of DDC-induced liver injury/regeneration in mice and reproducibility between hepatocyte-specific RNA-seq experiments.
a,b, Alanine aminotransferase (ALT) (a) and aspartate aminotransferase (AST) (b) levels in the serum of chow-fed mice (n = 6 animals) or 4-week DDC-fed mice (n = 3 animals). For a and b, parametric t test, unpaired with Welch’s correction, two-sided. The center value represents the mean, and error bars show s.d. c, Representative histology and immunofluorescence of chow- and DDC-fed mouse liver sections. First column: black arrows point to porphyrrin plugs, hepatic necrosis, and inflammatory cells. PV, portal vein. Second column: white arrows point to ductular hyperplasia (KRT19, red). Third column: white arrows point to proliferating hepatocyte nuclei co-stained for Hnf4-a (green) and pHistone3 (red). These experiments repeated with n = 3 animals in each condition. d, TPM (n = 2 biologically independent animals/condition) of cell-type-specific markers from hepatocytes and whole liver RNA-seq for chow- and DDC-fed conditions. e, Relative mRNA expression (qPCR) of hepatocyte- and non-parenchymal cell (NPC)-specific gene markers in hepatocytes isolated from embryonic day 18 (E18) mouse livers. f–h, Scatterplots of TPM values derived from paired-end RNA-seq biological replicate experiments. The number of genes with TPM >1 and Pearson correlation coefficients are indicated.
Supplementary Figure 2 Overlap of mRNA abundance and gene ontologies between development and regeneration clusters.
a, Histogram of the percentile score distribution of E18-specific (left; bottom 50% adult, top 50% E18) and adult chow-specific (right; top 50% adult, bottom 50% E18) genes. b, Gene Ontology (GO) terms of upregulated and downregulated genes in development and regeneration. c–e, Enrichment map of GO categories for each cluster are shown below (P < 0.05). The node size is proportionate to the number of genes related to each GO category, and the thickness of edges is proportionate to the number of shared genes between categories. For b–e, the sample size used was the same as in Fig. 1, n = 2 biologically independent animals/condition. Gene ontology enrichment analysis was performed using DAVID, with the expressed gene set as background and adjusting P values to account for multiple testing.
Supplementary Figure 3 Experimental approach and reproducibility of hepatocyte-specific in vivo polysome profiling from mouse livers.
a, Schematic description of the experimental procedure. Mouse livers were perfused with cycloheximide to stall ribosomes prior to collagenase digestion. The cytoplasmic lysate from purified hepatocytes was partitioned in a 15–45% sucrose gradient. A representative polysome profile is shown from adult mouse hepatocytes. RNA purified from different fractions was further processed to make sequencing libraries. b–i, Scatterplots of TPM values (biological replicates) derived from hepatocyte sequencing data for cytoplasmic (b,c), heavy polysome (d,e,), monosome (f,g), and light polysome (h,i) fractions prepared from chow- and DDC-fed mouse livers. Spearman correlation coefficients are indicated for each scatterplot. j, Scatterplot comparing ribosomal occupancies and gene expression changes in chow and DDC conditions for RNA-binding proteins and transcriptional factors. The sample size for b–j was n = 2 biologically independent animals/condition.
Supplementary Figure 4 Reversal of hepatocellular damage and regeneration response after DDC recovery.
a, Representative histology and immunofluorescence analysis of wild-type mouse liver sections on chow (n = 6 biologically independent animals) after 4 weeks of the DDC diet (n = 4) (injury phase) followed by 4 weeks of regular chow diet (recovery phase) (n = 5). First column: yellow arrows point to porphyrrin plugs and black arrows point to hepatic necrosis and inflammatory cells. PV, portal vein. Second column: white arrows point to proliferating hepatocyte nuclei co-stained for Hnf4-a (green) and pHistone3 (red) and orange arrows point to proliferating non-hepatocytes stained with pHistone3 only. 3rd column: White arrows point to ductular hyperplasia (KRT19, red). b–d, Quantifications of liver to body weight ratios (hepatosomatic index) (b), hepatocyte proliferation index (c), and ductular reaction (d) during the injury and recovery phases. In b–d, each point indicates the value for a single field of view in the section (n = 5 fields/animal). Data plots are mean ± s.d.; t test (two-tailed) with Welch’s correction was used to determine P values, and P < 0.05 was considered significant. e, Western blot showing hepatic ESRP2 protein levels after different days of DDC injury. TBP served as a loading control. The experiment was repeated independently three times; a representative image is shown.
Supplementary Figure 5 Generation of tetracycline-inducible hepatocyte-specific ESRP2 transgenic mice.
a, The TRE-ESRP2 construct expressing mouse ESRP2 cDNA with an N-terminal FLAG tag is driven by a tetracycline response element (TRE) and CMV minimal promoter. TRE-ESRP2 hemizygous transgenic mice were mated with ApoE-rtTA hemizygous transgenic mice to generate TRE-ESRP2; ApoE-rtTA bitransgenic mice as shown. Eight-week-old adult bitransgenic and littermate control (ApoE-rtTA) mice were fed doxycycline (Dox)-containing diet for 2 weeks to induce FLAG-ESRP2 expression specifically in hepatocytes. b, Western blotting against FLAG shows that exogenous ESRP2 in the liver is only expressed when Dox is present in the diet. Western blotting against ESRP2 shows the relative amount of induction compared to littermate controls. TBP was used as a loading control. c, FLAG and ESRP2 western blots showing regulated and dose-dependent hepatic ESRP2 protein expression after different Dox titrations in the diet. d, Western blot for FLAG and ESRP2 showing relative expression in the liver after induction with 0.5 g/kg Dox under chow and DDC injury conditions. All blots were repeated independently three times.
Supplementary Figure 6 RT–PCR validations and analyses of alternative splicing for the core Hippo pathway genes in WT, Esrp2 KO and ESRP2 overexpression livers under chow and DDC conditions.
a, Reciprocal regulation of Arhgf10l and Lsm14b alternative exons in Esrp2 KO and ESRP2 overexpression (OE) livers under chow-fed conditions. Skipping of these alternative exons after DDC-induced liver injury is rescued upon ESRP2 overexpression. The numeral after the gene name signifies the size of the alternative exon. The bands corresponding to (+) indicate exon inclusion and (–) indicate exon skipping. Percent spliced in (PSI) data are shown as the mean ± s.d.; n = 3. b, A simplified schematic of the Hippo signaling pathway. The core genes within the pathway that harbor ESRP2-regulated alternative exons (Nf2, Csnk1d, Yap1, and Tead1) are highlighted in orange. c, RT–PCR validation of ESRP2-mediated alternative splicing for the core Hippo pathway genes under chow- and DDC-induced liver injury conditions. PSI data are shown as the mean ± s.d.; n = 3. d,e, Western blotting for ESRP2 and TBP (d) and RT–PCR analysis of exon inclusion levels of the four Hippo pathway genes (e) 48 h after siRNA-based ESRP2 depletion in AML12 hepatocytes. PSI data are shown as the mean ± s.d.; n = 3. f, Map of upstream and downstream introns of Nf2, Csnk1d, Yap1 and Tead1 alternatively spliced exons showing occurrences of the ESRP2 binding motif (Fig. 6a).
Supplementary Figure 7 Targeting design and validation of antisense oligonucleotide specificity in switching the splicing pattern of core Hippo pathway genes.
a, Schematic of antisense oligonucleotide (ASO) design for targeting the 45nt Nf2, 63nt Csnk1d, 48nt Yap1 and 12nt Tead1 alternative exons. The alternative exons are shown in black, and the upstream and downstream constitutive exons are shown in grey colors. Red comb indicates the site of ASO complementarity to the pre-mRNA. ss: splice site. b, RT-PCR analyses of three unrelated ESRP2 regulated alternative exons in AML12 cells show no difference following treatment with control or a mixture of Hippo targeting ASOs demonstrating their target specificity. Percent spliced in (PSI) data are mean ± s.d.; n = 3. c, Relative mRNA levels of indicated Hippo pathway genes Nf2, Csnk1d, Yap1, and Tead1 normalized to Tbp (qPCR) following treatment with control or a mixture of Hippo targeting ASOs. Data are mean ± s.d.; n = 3. d, ChIP-qPCR for Hnf4a and Birc5 promoter regions spanning TEAD binding sites. e, Western blot for YAP1 and TBP post control and Yap1 targeting siRNA treatment for 24 hours in AML12 cells. siRNA knockdown repeated independently confirmed twice. f, MTT assay for AML12 cells under various conditions. Each point represents a biological replicate (n = 5). All data plots are mean ± s.d.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 and Supplementary Tables 1–3
Supplementary Dataset 1
Uncropped blot images
Supplementary Dataset 2
Bioinformatics source data
Supplementary Dataset 3
Experiment source data
Rights and permissions
About this article
Cite this article
Bangru, S., Arif, W., Seimetz, J. et al. Alternative splicing rewires Hippo signaling pathway in hepatocytes to promote liver regeneration. Nat Struct Mol Biol 25, 928–939 (2018). https://doi.org/10.1038/s41594-018-0129-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-018-0129-2