Abstract
The presence of active neurogenic niches in adult humans is controversial. We focused attention to the human olfactory neuroepithelium, an extracranial site supplying input to the olfactory bulbs of the brain. Using single-cell RNA sequencing analyzing 28,726 cells, we identified neural stem cell and neural progenitor cell pools and neurons. Additionally, we detailed the expression of 140 olfactory receptors. These data from the olfactory neuroepithelium niche provide evidence that neuron production may continue for decades in humans.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Code availability
The code used for the olfactory receptor analysis is available at https://github.com/harbourlab/OR_SC_analysis.
References
Hinds, J. W., Hinds, P. L. & McNelly, N. A. An autoradiographic study of the mouse olfactory epithelium: evidence for long-lived receptors. Anat. Rec. 210, 375–383 (1984).
Schwob, J. E., Szumowski, K. E. & Stasky, A. A. Olfactory sensory neurons are trophically dependent on the olfactory bulb for their prolonged survival. J. Neurosci. 12, 3896–3919 (1992).
Leung, C. T., Coulombe, P. A. & Reed, R. R. Contribution of olfactory neural stem cells to tissue maintenance and regeneration. Nat. Neurosci. 10, 720–726 (2007).
Goldstein, B. J. et al. Adult c-Kit+ progenitor cells are necessary for maintenance and regeneration of olfactory neurons. J. Comp. Neurol. 523, 15–31 (2015).
Fletcher, R. B. et al. Deconstructing olfactory stem cell trajectories at single-cell resolution. Cell Stem Cell 20, 817–830.e8 (2017).
Brann, J. H. & Firestein, S. J. A lifetime of neurogenesis in the olfactory system. Front. Neurosci. 8, 182 (2014).
Hahn, C. G. et al. In vivo and in vitro neurogenesis in human olfactory epithelium. J. Comp. Neurol. 483, 154–163 (2005).
Holbrook, E. H., Wu, E., Curry, W. T., Lin, D. T. & Schwob, J. E. Immunohistochemical characterization of human olfactory tissue. Laryngoscope 121, 1687–1701 (2011).
Graziadei, P. P., Karlan, M. S., Graziadei, G. A. & Bernstein, J. J. Neurogenesis of sensory neurons in the primate olfactory system after section of the fila olfactoria. Brain Res. 186, 289–300 (1980).
Becht, E. et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38–44 (2019).
van der Maaten, L. & Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008).
Corada, M. et al. Sox17 is indispensable for acquisition and maintenance of arterial identity. Nat. Commun. 4, 2609 (2013).
Chen, M., Reed, R. R. & Lane, A. P. Acute inflammation regulates neuroregeneration through the NF-κB pathway in olfactory epithelium. Proc. Natl Acad. Sci. USA 114, 8089–8094 (2017).
Schwob, J. E., Youngentob, S. L. & Mezza, R. C. Reconstitution of the rat olfactory epithelium after methyl bromide-induced lesion. J. Comp. Neurol. 359, 15–37 (1995).
Monahan, K., Horta, A. & Lomvardas, S. LHX2- and LDB1-mediated trans interactions regulate olfactory receptor choice. Nature 565, 448–453 (2019).
Hirota, J. & Mombaerts, P. The LIM-homeodomain protein Lhx2 is required for complete development of mouse olfactory sensory neurons. Proc. Natl Acad. Sci. USA 101, 8751–8755 (2004).
Wallrabenstein, I. et al. The smelling of Hedione results in sex-differentiated human brain activity. Neuroimage 113, 365–373 (2015).
Lyons, D. B. et al. An epigenetic trap stabilizes singular olfactory receptor expression. Cell 154, 325–336 (2013).
Buck, L. & Axel, R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell 65, 175–187 (1991).
Chess, A., Simon, I., Cedar, H. & Axel, R. Allelic inactivation regulates olfactory receptor gene expression. Cell 78, 823–834 (1994).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
McInnes, L., Healy, J., Saul, N. & Grossberger, L. UMAP: uniform manifold approximation and projection. J. Open Source Softw. 3, 861 (2018).
Blondel, V. D., Guillaume, J.-L., Lambiotte, R. & Lefebvre, E. Fast unfolding of communities in large networks. J. Stat. Mech. 2008, P10008 (2008).
Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016).
Gu, Z., Eils, R. & Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016).
Korotkevich, G., Sukhov, V. & Sergushichev, A. Fast gene set enrichment analysis. Preprint at bioRxiv https://doi.org/10.1101/060012 (2019).
Ligtenberg, W. reactome.db: a set of annotation maps for reactome. R package version 1.68.0 http://bioconductor.org/packages/release/data/annotation/html/reactome.db.html (2019).
Fabregat, A. et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 46, D649–D655 (2018).
Goldstein, B. J. et al. Contribution of Polycomb group proteins to olfactory basal stem cell self-renewal in a novel c-KIT+ culture model and in vivo. Development 143, 4394–4404 (2016).
Acknowledgements
We are grateful to the patients who generously contributed samples for this research. We acknowledge the assistance of the Onco-Genomics Shared Resource at the Sylvester Comprehensive Cancer Center and the University of Miami Center for Computational Science. This work was supported by funding from National Institutes of Health grant nos. DC013556 (B.J.G), DC016859 (B.J.G) and CA125970 (J.W.H.), the Triological Society/American College of Surgeons (B.J.G.), the University of Miami Sheila and David Fuente Graduate Program in Cancer Biology (M.A.D.) and the Center for Computational Science Fellowship (M.A.D.). The Sylvester Comprehensive Cancer Center also received funding from the National Cancer Institute Core Support Grant no. P30CA240139.
Author information
Authors and Affiliations
Contributions
M.A.D. and Stefan Kurtenbach analyzed and interpreted the data and wrote the manuscript. B.J.G. designed and led the project, performed the experiments, interpreted the data and wrote the manuscript. Z.B.S. provided the clinical samples and interpreted the data. J.W.H. designed the experiments and interpreted the data. R.C., G.M.G and Sarah Kurtenbach performed the experiments and interpreted the data. H.M. interpreted the data and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
J.W.H. is the inventor of intellectual property related to prognostic testing for uveal melanoma. He is a paid consultant for Castle Biosciences, a licensee of this intellectual property and receives royalties from its commercialization. The other authors do not have any potential competing interests.
Additional information
Peer review information Nature Neuroscience thanks Thomas Berger, Sandrine Thuret and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 DotPlot visualization listing scRNA-seq clusters.
a, Cell phenotypes listed on y-axis, showing unbiased gene expression for the top 8 genes per cluster identified by log Fold Change; genes (features) are listed along the x-axis. Dot size reflects percentage of cells in a cluster expressing each gene; dot color reflects expression level (as indicated on legend). The plot depicts clusters from 28,726 combined olfactory and respiratory mucosal cells, n = 4 patients. b, DotPlot visualization of the Heatmap shown in Fig. 1d. The plot depicts clusters from 28,726 combined olfactory and respiratory mucosal cells, n = 4 patients. c, Histogram showing the neuronal lineage cell types captured by scRNA-seq, as a percentage of the total cells analyzed per patient; see Fig. 1c.
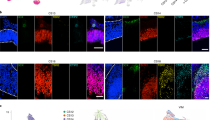
Extended Data Fig. 2 Additional human immunohistochemistry of basal cell populations.
Co-staining for SOX2, Ki67 or the HBC marker TP63. Proliferative activity has been used as a hallmark of the globose basal cell (GBC) phenotype. We reasoned that, although some proliferating cells in the olfactory epithelium (OE) might be immune or inflammatory cells, proliferative cells within the GBC layers of the OE that are SOX2+/Ki67+/TP63- would be categorized as GBCs. a, Sustentacular cell nuclei at the top of the OE are SOX2-bright; and horizontal basal cells (HBCs) and a subset of GBCs are SOX2+, although less intensely. Arrow marks a SOX2+/KI67+ cell among the proliferative KI67+ basal region, consistent with the GBC phenotype. b, SOX2 co-localizes with TP63 in HBCs; arrows mark Sox2+/TP63- GBCs. c, TP63 + HBCs are mitotically quiescent, while many GBCs are actively proliferating, often in scattered cell clusters. Arrow marks a cluster of KI67+/TP63- basal cells. Dashed line indicates basal lamina. Immunostaining for a-c was conducted in triplicate with similar results. Scale bar, 50 µm.
Extended Data Fig. 3 Additional human immunohistochemistry (IHC) of immature and mature olfactory sensory neurons (OSN) populations.
a, Co-staining for LHX2 and OMP demonstrates many LHX2+/OMP- neurons, distributed in deeper layers of the OE, which are immature OSNs; OMP is a marker for fully differentiated OSNs, while LHX2 expression in differentiating OSNs orchestrates OR receptor expression. b, DCX was identified by scRNA-seq here as enriched in immature OSNs (see Fig. 2a). IHC confirms scattered DCX+ neuronal somata and dendrites in the OE. c, Similarly, the bHLH transcription factor OLIG2 was identified to be enriched in immature OSNs; IHC confirms nuclear expression in the deeper OSN layers of OE tissue. Dashed line indicates basal lamina. Immunostaining for a–c was conducted in triplicate with similar results. Scale bar, 25 µm.
Extended Data Fig. 4 Analysis of immune cell populations.
Feature plots indicate expression of inflammatory cell markers in human nasal biopsy samples. UMAP clustering identifies lymphocyte populations; a, Cytotoxic T cell markers co-localize including CD8A, PRF1, GZMA and GZMB. The plot depicts clusters from 28,726 combined olfactory and respiratory mucosal cells, n = 4 patients. b, Within the monocyte/macrophage populations (CD14+, CD68+ cells), markers for activated M2 macrophages, such as CD163 and IL10, are indicated. The plot depicts clusters from 28,726 combined olfactory and respiratory mucosal cells, n = 4 patients. c, DotPlot visualization of additional immune cell gene expression from combined aggregate samples. Cell cluster identity is listed on the y-axis, genes (Features) are listed on x-axis. The plot depicts clusters from 28,726 combined olfactory and respiratory mucosal cells, n = 4 patients. d, Immune cell DotPlots showing the contribution of cell types and gene expression patterns by individual patient sample. The plot depicts clusters from 28,726 combined olfactory and respiratory mucosal cells, n = 4 patients.
Extended Data Fig. 6 Gene set enrichment analysis on differential expression data from selected cell clusters.
The top 50 Reactome pathways ranked by adjusted P value were plotted in the visualization. a, mOSNs versus GBCs; many top terms involve neuronal, transduction and synapse function. The differential expression was calculated in Seurat from 222 mOSNs and 115 GBCs, n = 4 patients. The default two-sided enrichment p-value with Benjamini–Hochberg correction from the fgsea package was utilized. b, GBCs versus olfactory HBCs; top terms include cell cycle or neurogenesis functions. The differential expression was calculated in Seurat from 115 GBCs and 2,182 olfactory HBCs, n = 4 patients. The default two-sided enrichment p-value with Benjamini–Hochberg correction from the fgsea package was utilized.
Extended Data Fig. 7 OR gene expression in human olfactory neurons.
a, Range of gene expression in our datasets (binned to 0.01). We identified 4.80 × 107 observations (gene expression measurements) expressing > 0. Genes with no expression ( = 0, n = 532063621) were excluded in this plot. The distribution plot shows that choosing a cutoff of 0.5 (red vertical dotted line). b, Doublet analysis. Box plots depicting the number of UMIs (“nCount_RNA”, left plot), and genes (“nFeature_RNA”, right plot) in immature and mature neurons expressing 1 or 2 ORs.
Extended Data Fig. 8 Principal component determination analysis.
The 4-patient combined data set was analyzed in Seurat to explore principal components (PCs) contributing to heterogeneity, and to determine an appropriate PC selection. a, Using the JackStraw approach, approximately 100 PCs had low a p-value. The plot depicts PC calculated from from 28,726 combined olfactory and respiratory mucosal cells, n = 4 patients. The jackstraw test implemented in Seurat was used to calculate p-values of PCs. b, To select a suitable number of PCs for downstream analysis, we used the elbow plot heuristic approach, indicating that beyond 20–30 PCs, very little additional variation is explained. Therefore, for downstream analysis we chose to include 30 PCs.
Extended Data Fig. 9 scRNA-seq quality control plots.
a, Number of genes (nFeature_RNA) per cluster. The plot depicts clusters from 28,726 combined olfactory and respiratory mucosal cells, n = 4 patients. b, Number of UMIs (nCount_RNA) per cluster. The plot depicts clusters from 28,726 combined olfactory and respiratory mucosal cells, n = 4 patients. Cluster cell type identities are listed along the x-axis. Violin plot widths are proportional to the density of the distribution.
Supplementary information
Supplementary Information
Supplementary Tables 1 and 11.
Supplementary Tables
Supplementary Tables 2–10.
Source data
Source Data Fig. 2
Statistical Source Data
Source Data Fig. 3
Statistical Source Data
Source Data Extended Data Fig. 1
Statistical Source Data
Source Data Extended Data Fig. 7
Statistical Source Data
Rights and permissions
About this article
Cite this article
Durante, M.A., Kurtenbach, S., Sargi, Z.B. et al. Single-cell analysis of olfactory neurogenesis and differentiation in adult humans. Nat Neurosci 23, 323–326 (2020). https://doi.org/10.1038/s41593-020-0587-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-020-0587-9
This article is cited by
-
Distinct patterns of cell adhesion, migration, and morphology in olfactory neuroepithelium cells of bipolar disorder patients
Molecular Medicine (2024)
-
Ambient air pollution undermines chemosensory sensitivity – a global perspective
Scientific Reports (2024)
-
Olfactory immunology: the missing piece in airway and CNS defence
Nature Reviews Immunology (2024)
-
Type 2 and Non-type 2 Inflammation in the Upper Airways: Cellular and Molecular Alterations in Olfactory Neuroepithelium Cell Populations
Current Allergy and Asthma Reports (2024)
-
Y-chromosome in the olfactory neuroepithelium as a potential biomarker of depression in women with male offspring: an exploratory study
Molecular and Cellular Biochemistry (2024)