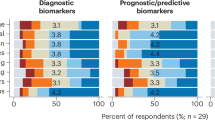
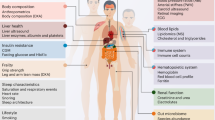
Abstract
The search for biomarkers that quantify biological aging (particularly ‘omic’-based biomarkers) has intensified in recent years. Such biomarkers could predict aging-related outcomes and could serve as surrogate endpoints for the evaluation of interventions promoting healthy aging and longevity. However, no consensus exists on how biomarkers of aging should be validated before their translation to the clinic. Here, we review current efforts to evaluate the predictive validity of omic biomarkers of aging in population studies, discuss challenges in comparability and generalizability and provide recommendations to facilitate future validation of biomarkers of aging. Finally, we discuss how systematic validation can accelerate clinical translation of biomarkers of aging and their use in gerotherapeutic clinical trials.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Moqri, M. et al. Biomarkers of aging for the identification and evaluation of longevity interventions. Cell 186, 3758–3775 (2023).
Moqri, M. et al. PRC2 clock: a universal epigenetic biomarker of aging and rejuvenation. Preprint at bioRxiv https://doi.org/10.1101/2022.06.03.494609 (2022).
Poganik, J. R. et al. Biological age is increased by stress and restored upon recovery. Cell Metab. 35, 807–820 (2023).
Lu, A. T. et al. DNA methylation GrimAge version 2. Aging 14, 9484–9549 (2022).
Tanaka, T. et al. Plasma proteomic biomarker signature of age predicts health and life span. eLife 9, e61073 (2020).
Balasubramanian, R. et al. Metabolomic profiles associated with all-cause mortality in the Women’s Health Initiative. Int. J. Epidemiol. 49, 289–300 (2020).
Sarkar, T. J. et al. Transient non-integrative expression of nuclear reprogramming factors promotes multifaceted amelioration of aging in human cells. Nat. Commun. 11, 1545 (2020).
Lee, J. W. et al. Fit-for-purpose method development and validation for successful biomarker measurement. Pharm. Res. 23, 312–328 (2006).
Wagner, J. A. Overview of biomarkers and surrogate endpoints in drug development. Dis. Markers 18, 41–46 (2002).
Hunter, D. J. et al. A pathway and approach to biomarker validation and qualification for osteoarthritis clinical trials. Curr. Drug Targets 11, 536–545 (2010).
Bortz, J. et al. Biological age estimation using circulating blood biomarkers. Commun. Biol. 6, 1089 (2023).
Lehallier, B. et al. Undulating changes in human plasma proteome profiles across the lifespan. Nat. Med. 25, 1843–1850 (2019).
Ahadi, S. et al. Personal aging markers and ageotypes revealed by deep longitudinal profiling. Nat. Med. 26, 83–90 (2020).
Alpert, A. et al. A clinically meaningful metric of immune age derived from high-dimensional longitudinal monitoring. Nat. Med. 25, 487–495 (2019).
Rutledge, J., Oh, H. & Wyss-Coray, T. Measuring biological age using omics data. Nat. Rev. Genet. 23, 715–727 (2022).
Kudryashova, K. S., Burka, K., Kulaga, A. Y., Vorobyeva, N. S. & Kennedy, B. K. Aging biomarkers: from functional tests to multi‐omics approaches. Proteomics 20, 1900408 (2020).
Committee on the Review of Omics-Based Tests for Predicting Patient Outcomes in Clinical Trials; Board on Health Care Services; Board on Health Sciences Policy; & Institute of Medicine. Evolution of Translational Omics: Lessons Learned and the Path Forward (eds Micheel, C. M. et al.) (National Academies Press, 2012).
Ying, K. et al. ClockBase: a comprehensive platform for biological age profiling in human and mouse. Preprint at bioRxiv https://doi.org/10.1101/2023.02.28.530532 (2023).
Ying, K. et al. Causality-enriched epigenetic age uncouples damage and adaptation. Nat. Aging https://doi.org/10.1038/s43587-023-00557-0 (2024).
Belsky, D. W. et al. DunedinPACE, a DNA methylation biomarker of the pace of aging. eLife 11, e73420 (2022).
Bautmans, I. et al. WHO working definition of vitality capacity for healthy longevity monitoring. Lancet Healthy Longev. 3, e789–e796 (2022).
Lara, J. et al. A proposed panel of biomarkers of healthy ageing. BMC Med. 13, 222 (2015).
Crimmins, E. M., Thyagarajan, B., Levine, M. E., Weir, D. R. & Faul, J. Associations of age, sex, race/ethnicity, and education with 13 epigenetic clocks in a nationally representative U.S. sample: the Health and Retirement Study. J. Gerontol. A Biol. Sci. Med. Sci. 76, 1117–1123 (2021).
Faul, J. D. et al. Epigenetic-based age acceleration in a representative sample of older Americans: associations with aging-related morbidity and mortality. Proc. Natl Acad. Sci. USA 120, e2215840120 (2023).
Lu, A. T. et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging 11, 303–327 (2019).
Gadd, D. A. et al. Blood protein levels predict leading incident diseases and mortality in UK Biobank. Preprint at medRxiv https://doi.org/10.1101/2023.05.01.23288879 (2023).
Tian, Y. E. et al. Heterogeneous aging across multiple organ systems and prediction of chronic disease and mortality. Nat. Med. 29, 1221–1231 (2023).
Buergel, T. et al. Metabolomic profiles predict individual multidisease outcomes. Nat. Med. 28, 2309–2320 (2022).
Raghavachari, N., Wilmot, B. & Dutta, C. Optimizing translational research for exceptional health and life span: a systematic narrative of studies to identify translatable therapeutic target(s) for exceptional health span in humans. J. Gerontol. A Biol. Sci. Med. Sci. 77, 2272–2280 (2022).
Kurki, M. I. et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature 613, 508–518 (2023).
Nagai, A. et al. Overview of the BioBank Japan Project: study design and profile. J. Epidemiol. 27, S2–S8 (2017).
Boutin, N. T. et al. The evolution of a large biobank at Mass General Brigham. J. Pers. Med. 12, 1323 (2022).
Huan, T. et al. Integrative analysis of clinical and epigenetic biomarkers of mortality. Aging Cell 21, e13608 (2022).
Eiriksdottir, T. et al. Predicting the probability of death using proteomics. Commun. Biol. 4, 758 (2021).
Deelen, J. et al. A metabolic profile of all-cause mortality risk identified in an observational study of 44,168 individuals. Nat. Commun. 10, 3346 (2019).
Kuiper, L. M. et al. Epigenetic and metabolomic biomarkers for biological age: a comparative analysis of mortality and frailty risk. J. Gerontol. A Biol. Sci. Med. Sci. 78, 1753–1762 (2023).
Wang, C. et al. DNA methylation-based biomarkers of age acceleration and all-cause death, myocardial infarction, stroke, and cancer in two cohorts: the NAS, and KORA F4. eBioMedicine 63, 103151 (2021).
Evans, M. K. et al. Healthy Aging in Neighborhoods of Diversity across the Life Span (HANDLS): overcoming barriers to implementing a longitudinal, epidemiologic, urban study of health, race, and socioeconomic status. Ethn. Dis. 20, 267–275 (2010).
Olivia, S. et al. Comparison of DNA methylation measured by Illumina 450K and EPIC BeadChips in blood of newborns and 14-year-old children. Epigenetics 13, 655–664 (2018).
Koistinen, V. et al. Towards a Rosetta stone for metabolomics: recommendations to overcome inconsistent metabolite nomenclature. Nat. Metab. 5, 351–354 (2023).
Fahy, E. & Subramaniam, S. RefMet: a reference nomenclature for metabolomics. Nat. Methods 17, 1173–1174 (2020).
Yu, B. et al. The Consortium of Metabolomics Studies (COMETS): metabolomics in 47 prospective cohort studies. Am. J. Epidemiol. 188, 991–1012 (2019).
Psaty, B. M. et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ. Cardiovasc. Genet. 2, 73–80 (2009).
Keane, T. & Glass, L. CINECA: Common Infrastructure for National Cohorts in Europe, Canada, and Africa — Kick Off Report. Zenodo https://doi.org/10.5281/zenodo.3908145 (2019).
Taliun, D. et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature 590, 290–299 (2021).
Linsen, L. et al. Raising to the challenge: building a federated biobank to accelerate translational research—the University Biobank Limburg. Front. Med. 6, 224 (2019).
Kaye, J. et al. Access governance for biobanks: the case of the BioSHaRE-EU cohorts. Biopreserv. Biobank. 14, 201–206 (2016).
Cummings, S. R. & Kritchevsky, S. B. Endpoints for geroscience clinical trials: health outcomes, biomarkers, and biologic age. GeroScience 44, 2925–2931 (2022).
Kaeberlein, M. How healthy is the healthspan concept? GeroScience 40, 361–364 (2018).
Okada, D., Cheng, J. H., Zheng, C., Kumaki, T. & Yamada, R. Data-driven identification and classification of nonlinear aging patterns reveals the landscape of associations between DNA methylation and aging. Hum. Genomics 17, 8 (2023).
Higgins-Chen, A. T. et al. A computational solution for bolstering reliability of epigenetic clocks: implications for clinical trials and longitudinal tracking. Nat. Aging 2, 644–661 (2022).
Tomusiak, A. et al. Development of a novel epigenetic clock resistant to changes in immune cell composition. Preprint at bioRxiv https://doi.org/10.1101/2023.03.01.530561 (2023).
Fang, F. et al. Evaluation of pediatric epigenetic clocks across multiple tissues. Clin. Epigenetics 15, 142 (2023).
Johnson, N. D. et al. Non-linear patterns in age-related DNA methylation may reflect CD4+ T cell differentiation. Epigenetics 12, 492–503 (2017).
Zhou, W. et al. SeSAMe: Reducing artifactual detection of DNA methylation by Infinium BeadChips in genomic deletions. Nucleic Acids Res. 46, 20 (2018).
Emmanuel, T. et al. A survey on missing data in machine learning. J. Big Data 8, 140 (2021).
Li, P., Stuart, E. A. & Allison, D. B. Multiple imputation: a flexible tool for handling missing data. JAMA 314, 1966–1967 (2015).
Paul, Y. et al. Considerations for normalization of DNA methylation data by Illumina 450K BeadChip assay in population studies. Epigenetics 8, 1141–1152 (2013).
Bizzarri, D., Reinders, M. J. T., Beekman, M., Slagboom, P. E. & van den Akker, E. B. MiMIR: R-shiny application to infer risk factors and endpoints from Nightingale Health’s 1H-NMR metabolomics data. Bioinformatics 38, 3847–3849 (2022).
Kwon, D. & Belsky, D. W. A toolkit for quantification of biological age from blood chemistry and organ function test data: BioAge. GeroScience 43, 2795–2808 (2021).
Ying, K. et al. Biolearn, an open-source library for biomarkers of aging. Preprint at bioRxiv https://doi.org/10.1101/2023.12.02.569722 (2023).
Thrush, K. L., Higgins-Chen, A. T., Liu, Z. & Levine, M. E. R methylCIPHER: a methylation clock investigational package for hypothesis-driven evaluation & research. Preprint at bioRxiv https://doi.org/10.1101/2022.07.13.499978 (2022).
Pun, F. W. et al. Hallmarks of aging-based dual-purpose disease and age-associated targets predicted using PandaOmics AI-powered discovery engine. Aging 14, 2475–2506 (2022).
Manning, A. K. et al. NHLBI BioData Catalyst and the future of cloud computing. Genet. Epidemiol. 45, 774–775 (2021).
Li, T., Sahu, A. K., Talwalkar, A. & Smith, V. Federated learning: challenges, methods, and future directions. IEEE Signal Process. Mag. 37, 50–60 (2020).
Fortier, I. et al. Maelstrom Research guidelines for rigorous retrospective data harmonization. Int. J. Epidemiol. 46, 103–105 (2017).
Wilkinson, M. et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 3, 160018 (2016).
Tang, M. X. et al. The APOE-ε4 allele and the risk of Alzheimer disease among African Americans, whites, and Hispanics. JAMA 279, 751–755 (1998).
Benjamin, C. T. et al. Apolipoprotein E4 is associated with improved cognitive function in Amazonian forager–horticulturalists with a high parasite burden. FASEB J. 31, 1251–1767 (2017).
Privé, F. et al. Portability of 245 polygenic scores when derived from the UK Biobank and applied to 9 ancestry groups from the same cohort. Am. J. Hum. Genet. 109, 12–23 (2022).
Graf, G. H. et al. Testing Black–white disparities in biological aging among older adults in the United States: analysis of DNA-methylation and blood-chemistry methods. Am. J. Epidemiol. 191, 613–625 (2022).
Liu, Z. et al. A new aging measure captures morbidity and mortality risk across diverse subpopulations from NHANES IV: a cohort study. PLoS Med. 15, e1002718 (2018).
Vandenbroucke, J. P. et al. Strengthening the Reporting of Observational Studies in Epidemiology (STROBE): explanation and elaboration. PLoS Med. 4, e297 (2007).
Rodés, B., Cadiñanos, J., Esteban-Cantos, A., Rodríguez-Centeno, J. & Arribas, J. R. Ageing with HIV: challenges and biomarkers. eBioMedicine 77, 103896 (2022).
Cohen, A. A. et al. Detection of a novel, integrative aging process suggests complex physiological integration. PLoS ONE 10, e0116489 (2015).
Ferrucci, L. & Kohanski, R. Better care for older patients with complex multimorbidity and frailty: a call to action. Lancet Healthy Longev. 3, e581–e583 (2022).
Baker, G. T. & Sprott, R. L. Biomarkers of aging. Exp. Gerontol. 23, 223–239 (1988).
Levine, M. E. et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging 10, 573–591 (2018).
Sebastiani, P. et al. Biomarker signatures of aging. Aging Cell 16, 329–338 (2017).
Mamoshina, P. et al. Population specific biomarkers of human aging: a big data study using South Korean, Canadian, and Eastern European patient populations. J. Gerontol. A Biol. Sci. Med. Sci. 73, 1482–1490 (2018).
Zhang, Y. et al. DNA methylation signatures in peripheral blood strongly predict all-cause mortality. Nat. Commun. 8, 14617 (2017).
Bernabeu, E. et al. Refining epigenetic prediction of chronological and biological age. Genome Med. 15, 12 (2023).
van den Akker, E. B. et al. Metabolic age based on the BBMRI-NL 1H-NMR metabolomics repository as biomarker of age-related disease. Circ. Genom. Precis. Med. 13, 541–547 (2020).
Li, X. et al. Longitudinal trajectories, correlations and mortality associations of nine biological ages across 20-years follow-up. eLife 9, e51507 (2020).
Hillary, R. F. et al. Epigenetic measures of ageing predict the prevalence and incidence of leading causes of death and disease burden. Clin. Epigenet. 12, 115 (2020).
McCrory, C. et al. GrimAge outperforms other epigenetic clocks in the prediction of age-related clinical phenotypes and all-cause mortality. J. Gerontol. A Biol. Sci. Med. Sci. 76, 741–749 (2021).
Tiina, F. et al. Does the epigenetic clock GrimAge predict mortality independent of genetic influences: an 18 year follow-up study in older female twin pairs. Clin. Epigenetics 13, 128 (2021).
Acknowledgements
This work was supported in part by the Intramural Research Program of the National Institute on Aging, NIH, and grants from the National Institute on Aging and Hevolution Foundation. D.P.K. was supported by a grant from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (R01AR041398). We are very grateful to D.M. Wilson III for many suggestions during the writing of this paper. We express our gratitude to all members of the Biomarkers of Aging Consortium Roadmap Group (https://www.agingconsortium.org/) for their fruitful discussions that helped to define the scope and direction of this work. The list of authors reflects Roadmap Group members who directly contributed to or refined the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
M.M., V.S., M.P.S. and V.N.G. have filed a patent on measuring cellular aging. C.H. is also affiliated with the Institute for Biomedical Aging Research, Universität Innsbruck, Austria and is an honorary research fellow at the Department of Women’s Cancer, EGA Institute for Women’s Health, University College London. C.H. is a shareholder of Sola Diagnostics and is named as an inventor on a patent on an epigenetic clock indicative of breast cancer risk. J.N.J. is also affiliated with the Sticht Center for Healthy Aging and Alzheimer’s Prevention, Wake Forest University School of Medicine and the XPRIZE Foundation. J.N.J. serves on the advisory board for the American Federation for Aging Research’s Finding Aging Biomarkers by Searching Existing Trials Initiative and the editorial board of the Journals of Gerontology Series A Biological Sciences, eLife and Experimental Gerontology. D.W.B. is also affiliated with the Child Brain Development Network, Canadian Institute for Advanced Research and SocioMed Research Nucleus and Universidad Mayor. D.W.B. is an inventor of DunedinPACE, a Duke University and University of Otago invention licensed to TruDiagnostic. A.T.H.-C. is an inventor of epigenetic clocks that are the subject of a provisional patent and have been licensed to TruDiagnostic. A.T.H.-C. has also received consulting fees from TruDiagnostic and FOXO Biosciences. B.H.C. owns stock in Illumina, the manufacturer of the DNA methylation arrays used in epigenetic biomarkers of aging, and is listed as a co-inventor on filed patents on commercial applications of epigenetic prediction models. A.A.C. is a founder, president and majority shareholder at Oken Health. R.E.M. has received a speaker fee from Illumina and is an advisor to the Epigenetic Clock Development Foundation and Optima Partners. M.W. is also affiliated with the Institute for Biomedical Aging Research, Universität Innsbruck. M.W. is a shareholder of Sola Diagnostics and is named as an inventor on a patent on an epigenetic clock indicative of breast cancer risk. K.F. is the CEO of BioAge Labs. P.O.F. is an employee and stakeholder of Gero. A.Z. is the founder and the CEO of Insilico Medicine, a clinical-stage generative AI and robotics biotechnology company specializing in aging research. N.B. is the scientific director of the American Federation for Aging Research, is on the board of the executive committee of the Longevity Biotech Association and is advisor on the board of the Academy for Health and Lifespan Research. D.P.K. has received a grant from Solarea Bio and royalties from Wolters Kluwer. D.P.K. sits on the scientific advisory boards of Solarea Bio, Pfizer, Radius Health and Reneo and has participated in the data safety monitoring board for the AgNovos Healthcare treatment trial. E.V. is a scientific cofounder of Napa Therapeutics and BHB Therapeutics, serves on the scientific advisory board of Seneque and is a named co-inventor on a patent relating to an epigenetic clock robust to cell composition changes. A.B.M. declares herself chief medical officer of NU and co-founder of Chi Longevity. V.S. is a cofounder, SAB chair and head of research of Turn Biotechnologies. M.P.S. is a cofounder and scientific advisor of Personalis, SensOmics, Qbio, January AI, Fodsel, Filtricine, Protos, RTHM, iollo, Marble Therapeutics, Crosshair Therapeutics and Mirvie. He is a scientific advisor of Jupiter, Neuvivo, Swaza and Mitrix. S. Horvath is a founder of the nonprofit Epigenetic Clock Development Foundation that licenses patents surrounding epigenetic clocks. The Regents of the University of California is the sole owner of a patent application directed at GrimAge and other epigenetic clocks for which S. Horvath is a named inventor.
Peer review
Peer review information
Nature Medicine thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Karen O’Leary, in collaboration with the Nature Medicine team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Note and Tables 1–3
Rights and permissions
About this article
Cite this article
Moqri, M., Herzog, C., Poganik, J.R. et al. Validation of biomarkers of aging. Nat Med 30, 360–372 (2024). https://doi.org/10.1038/s41591-023-02784-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-023-02784-9