Abstract
Effective pharmacotherapy for major depressive disorder remains a major challenge, as more than 30% of patients are resistant to the first line of treatment (selective serotonin reuptake inhibitors)1. Sub-anaesthetic doses of ketamine, a non-competitive N-methyl-d-aspartate receptor antagonist2,3, provide rapid and long-lasting antidepressant effects in these patients4,5,6, but the molecular mechanism of these effects remains unclear7,8. Ketamine has been proposed to exert its antidepressant effects through its metabolite (2R,6R)-hydroxynorketamine ((2R,6R)-HNK)9. The antidepressant effects of ketamine and (2R,6R)-HNK in rodents require activation of the mTORC1 kinase10,11. mTORC1 controls various neuronal functions12, particularly through cap-dependent initiation of mRNA translation via the phosphorylation and inactivation of eukaryotic initiation factor 4E-binding proteins (4E-BPs)13. Here we show that 4E-BP1 and 4E-BP2 are key effectors of the antidepressant activity of ketamine and (2R,6R)-HNK, and that ketamine-induced hippocampal synaptic plasticity depends on 4E-BP2 and, to a lesser extent, 4E-BP1. It has been hypothesized that ketamine activates mTORC1–4E-BP signalling in pyramidal excitatory cells of the cortex8,14. To test this hypothesis, we studied the behavioural response to ketamine and (2R,6R)-HNK in mice lacking 4E-BPs in either excitatory or inhibitory neurons. The antidepressant activity of the drugs is mediated by 4E-BP2 in excitatory neurons, and 4E-BP1 and 4E-BP2 in inhibitory neurons. Notably, genetic deletion of 4E-BP2 in inhibitory neurons induced a reduction in baseline immobility in the forced swim test, mimicking an antidepressant effect. Deletion of 4E-BP2 specifically in inhibitory neurons also prevented the ketamine-induced increase in hippocampal excitatory neurotransmission, and this effect concurred with the inability of ketamine to induce a long-lasting decrease in inhibitory neurotransmission. Overall, our data show that 4E-BPs are central to the antidepressant activity of ketamine.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data generated and analysed during this study are included in this published Article and its Supplementary Information files. Source data are provided with this paper.
References
Rush, A. J. et al. Acute and longer-term outcomes in depressed outpatients requiring one or several treatment steps: a STAR*D report. Am. J. Psychiatry 163, 1905–1917 (2006).
Hirota, K. & Lambert, D. G. Ketamine: its mechanism(s) of action and unusual clinical uses. Br. J. Anaesth. 77, 441–444 (1996).
Kashiwagi, K. et al. Channel blockers acting at N-methyl-d-aspartate receptors: differential effects of mutations in the vestibule and ion channel pore. Mol. Pharmacol. 61, 533–545 (2002).
Berman, R. M. et al. Antidepressant effects of ketamine in depressed patients. Biol. Psychiatry 47, 351–354 (2000).
Zarate, C. A., Jr et al. A randomized trial of an N-methyl-d-aspartate antagonist in treatment-resistant major depression. Arch. Gen. Psychiatry 63, 856–864 (2006).
Lapidus, K. A. et al. A randomized controlled trial of intranasal ketamine in major depressive disorder. Biol. Psychiatry 76, 970–976 (2014).
Zarate, C. A., Jr & Machado-Vieira, R. Ketamine: translating mechanistic discoveries into the next generation of glutamate modulators for mood disorders. Mol. Psychiatry 22, 324–327 (2017).
Workman, E. R., Niere, F. & Raab-Graham, K. F. Engaging homeostatic plasticity to treat depression. Mol. Psychiatry 23, 26–35 (2018).
Zanos, P. et al. NMDAR inhibition-independent antidepressant actions of ketamine metabolites. Nature 533, 481–486 (2016).
Li, N. et al. mTOR-dependent synapse formation underlies the rapid antidepressant effects of NMDA antagonists. Science 329, 959–964 (2010).
Fukumoto, K. et al. Activity-dependent brain-derived neurotrophic factor signaling is required for the antidepressant actions of (2R,6R)-hydroxynorketamine. Proc. Natl Acad. Sci. USA 116, 297–302 (2019).
Saxton, R. A. & Sabatini, D. M. mTOR signaling in growth, metabolism, and disease. Cell 169, 361–371 (2017).
Sonenberg, N. & Hinnebusch, A. G. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136, 731–745 (2009).
Miller, O. H., Moran, J. T. & Hall, B. J. Two cellular hypotheses explaining the initiation of ketamine’s antidepressant actions: direct inhibition and disinhibition. Neuropharmacology 100, 17–26 (2016).
Kavalali, E. T. & Monteggia, L. M. The ketamine metabolite 2R,6R-hydroxynorketamine blocks NMDA receptors and impacts downstream signaling linked to antidepressant effects. Neuropsychopharmacology 43, 221–222 (2018).
Suzuki, K., Nosyreva, E., Hunt, K. W., Kavalali, E. T. & Monteggia, L. M. Effects of a ketamine metabolite on synaptic NMDAR function. Nature 546, E1–E3 (2017).
Paul, R. K. et al. (R,S)-Ketamine metabolites (R,S)-norketamine and (2S,6S)-hydroxynorketamine increase the mammalian target of rapamycin function. Anesthesiology 121, 149–159 (2014).
Adaikkan, C., Taha, E., Barrera, I., David, O. & Rosenblum, K. Calcium/calmodulin-dependent protein kinase II and eukaryotic elongation factor 2 kinase pathways mediate the antidepressant action of ketamine. Biol. Psychiatry 84, 65–75 (2018).
Miller, O. H., Grabole, N., Wells, I. & Hall, B. Genome-wide translating mRNA analysis following ketamine reveals novel targets for antidepressant treatment. Preprint at https://doi.org/10.1101/254904 (2018).
Murrough, J. W. et al. Rapid and longer-term antidepressant effects of repeated ketamine infusions in treatment-resistant major depression. Biol. Psychiatry 74, 250–256 (2013).
Aguilar-Valles, A., Matta-Camacho, E. & Sonenberg, N. in The Oxford Handbook of Neuronal Protein Synthesis (ed. Wayne Sossin) (Oxford Univ. Press, 2018).
Banko, J. L. et al. The translation repressor 4E-BP2 is critical for eIF4F complex formation, synaptic plasticity, and memory in the hippocampus. J. Neurosci. 25, 9581–9590 (2005).
Autry, A. E. et al. NMDA receptor blockade at rest triggers rapid behavioural antidepressant responses. Nature 475, 91–95 (2011).
Gerhard, D. M. et al. GABA interneurons are the cellular trigger for ketamine’s rapid antidepressant actions. J. Clin. Invest. 130, 1336–1349 (2020).
Duman, R. S., Sanacora, G. & Krystal, J. H. Altered connectivity in depression: GABA and glutamate neurotransmitter deficits and reversal by novel treatments. Neuron 102, 75–90 (2019).
Ng, L. H. L. et al. Ketamine and selective activation of parvalbumin interneurons inhibit stress-induced dendritic spine elimination. Transl. Psychiatry 8, 272 (2018).
Widman, A. J. & McMahon, L. L. Disinhibition of CA1 pyramidal cells by low-dose ketamine and other antagonists with rapid antidepressant efficacy. Proc. Natl Acad. Sci. USA 115, E3007–E3016 (2018).
Moda-Sava, R. N. et al. Sustained rescue of prefrontal circuit dysfunction by antidepressant-induced spine formation. Science 364, eaat8078 (2019).
Yang, Y. et al. Ketamine blocks bursting in the lateral habenula to rapidly relieve depression. Nature 554, 317–322 (2018).
Bidinosti, M. et al. Postnatal deamidation of 4E-BP2 in brain enhances its association with raptor and alters kinetics of excitatory synaptic transmission. Mol. Cell 37, 797–808 (2010).
Gkogkas, C. G. et al. Autism-related deficits via dysregulated eIF4E-dependent translational control. Nature 493, 371–377 (2013).
Jernigan, C. S. et al. The mTOR signaling pathway in the prefrontal cortex is compromised in major depressive disorder. Prog. Neuropsychopharmacol. Biol. Psychiatry 35, 1774–1779 (2011).
Abdallah, C. et al. Rapamycin, an immunosuppressant and mTORC1 inhibitor, triples the antidepressant response rate of ketamine at 2 weeks following treatment: a double-blind, placebo-controlled, cross-over, randomized clinical trial. Preprint at https://doi.org/10.1101/500959 (2018).
Tsukiyama-Kohara, K. et al. Adipose tissue reduction in mice lacking the translational inhibitor 4E-BP1. Nat. Med. 7, 1128–1132 (2001).
Bodnoff, S. R., Suranyi-Cadotte, B., Aitken, D. H., Quirion, R. & Meaney, M. J. The effects of chronic antidepressant treatment in an animal model of anxiety. Psychopharmacology (Berl.) 95, 298–302 (1988).
Acknowledgements
We thank A. Sylvestre, A. Lafrance, I. Harvey and T. Degenhard for technical assistance. A.A.-V. was a recipient of FRQS (Fond Recherche Québec-Santé) and CIHR (Canadian Institutes for Health Research) postdoctoral fellowships. D.D.G. is a recipient of FRQS and CIHR postdoctoral fellowships. J.-C.L. is the recipient of the Canada Research Chair in Cellular and Molecular Neurophysiology. This work was supported by a CIHR Foundation grant to N. Sonenberg (FND-148423), and a CIHR project grant to J.-C.L. (PJT-153311).
Author information
Authors and Affiliations
Contributions
A.A.-V. designed the study, performed experiments, supervised students, analysed data and wrote the manuscript. D.D.G. performed behavioural experiments and data analyses. E.M.-C. performed western blot experiments, supervised students and edited the manuscript. M.J.E. performed electrophysiological experiments. A.K. performed electrophysiological experiments and data analyses. A.S. performed immunohistochemistry experiments and contributed to the writing of the manuscript. M.L.-C. and A.T.-B. contributed to behavioural tests. S.B. performed data analyses. G.M.R., S.S. and N. Salmaso contributed to experimental design, data analysis and manuscript editing. G.G., J.-C.L. and N. Sonenberg supervised students, designed experiments and contributed to writing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Kimberly Raab-Graham and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
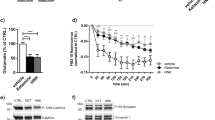
Extended Data Fig. 1 Expression of 4E-BP1 and 4E-BP2 in the prefrontal cortex and hippocampus.
a, Representative western blot analysis (from 3 independent experiments) of 4E-BP1 and 4E-BP2 in the prefrontal cortex and hippocampus of wild-type (Eif4ebp1/2+/+ +/+), 4E-BP1 knock out (Eif4ebp1−/−), 4E-BP2 knock out (Eif4ebp2−/−), and 4E-BP1/2 double knock out mice (Eif4ebp1/2−/−) (uncropped blots are presented in Supplementary Information). b, c, Representative confocal images of immunostaining for 4E-BP2 (green), CAMK2α (red)/GAD67 (magenta) in the hippocampus. The arrows indicate area showed in the insert, which is a higher magnification of the images. Insert scale bar: 10 μm. Brain sections were additionally counterstained with DAPI (blue). d, Representative confocal images of immunostaining for 4E-BP1 (green), GFAP (red) in the hippocampus. Brain sections were additionally counterstained with DAPI (blue). The white box indicates area shown in e. f, Representative confocal image of immunostaining for 4E-BP1 (green) and CD11b (magenta). Brain sections were additionally counterstained with DAPI (blue). All immunofluorescence experiments were repeated in 3 independent experiments.
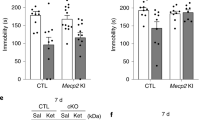
Extended Data Fig. 2 4E-BP1 and 4E-BP2 are required for the antidepressant effect of ketamine.
a, A group of Eif4ebp1/2+/+, Eif4ebp1−/−, and Eif4ebp2−/− mice were treated with saline or ketamine (IP, 10 mg kg−1) and tested 24 h after injection in the FST (2-way ANOVA and Fisher’s LSD, n = 11, 13, 11, 12, 4, 7 from left to right). b, Wild-type (Eif4ebp1/2+/+), Eif4ebp1−/− and Eif4ebp2−/− mice were treated with saline or ketamine (IP, 10 or 30 mg kg−1) and tested in the FST 6 days after treatment (2-way ANOVA and Fisher’s LSD, n = 13, 19, 7, 11, 10, 9, 16, 14, 6 from left to right). c, A different group of mice treated as in a was tested in the open field, and general activity was measured as distance travelled (2-way ANOVA, n = 8, 8, 8, 6, 6, 5, 6, 6, 7 from left to right). d, Latency to feed in the home cage was measured immediately after the NSF task in Eif4ebp1/2+/+ +/+, Eif4ebp1−/− and Eif4ebp2−/− mice, treated with saline or Ket (corresponding to Fig. 1b; 2-way ANOVA). e, Latency to feed in the home cage was also measured after the NSF task in the three mouse strains, following saline or (2R,6R)-HNK treatment (corresponding to Fig. 1c; 2-way ANOVA). f, In the same group of mice as in e (and Fig. 1c) average distance travelled during the NSF test was measured (expressed as an average per minute to correct for differential time spent in the arena; 2-way ANOVA). g, Mice of the three genotypes were also tested for immobility in the tail suspension test, 1 h after saline or Ket treatments (2-way ANOVA and Fisher’s LSD, n = 8, 9, 9, 10, 10, 7 from left to right). h, Eif4ebp1/2+/+ +/+, Eif4ebp1−/− and Eif4ebp2−/− mice were treated with fluoxetine (Fluox, IP, 3 mg kg−1, 0.5 h) and tested in the FST (2-way ANOVA and Fisher’s LSD, n = 8, 10, 9, 8, 6, 6 from left to right). Mean ± s.e.m.; * P < 0.05; ** P < 0.01 vs. Sal-treated mice of the same genotype.
Extended Data Fig. 3 Generation of Eif4ebp1 and Eif4ebp2 floxed genes in mice.
Schematic representation of the targeting construct used for the generation of Eif4ebp1 and Eif4ebp2 knockout mice. FRT (FLPase recognition target) and loxP sequences are indicated. Positive (PGK-neo) selection markers are indicated in the schemes on the Neomycin cassette (Neo, neomycin). Numbered grey boxes represent exons in the Eif4ebp1 and Eif4ebp2 genes. The Eif4ebp1 and Eif4ebp2 neomycin alleles were produced by homologous recombination. FLPase was used to generate the Eif4ebp1 and Eif4ebp2 floxed alleles. The genotyping PCR primers are indicated for each construct, as well as the length of the PCR product.
Extended Data Fig. 4 Effects of cell-specific mutation of Eif4ebp1 and Eif4ebp2 on home cage feeding, locomotion and forced swim test.
a, Latency to feed in the home cage was measured 1 h after saline or Ket (IP, 10 mg kg−1) treatments in control, mice lacking Eif4ebp1 in CAMK2α+ (Eif4ebp1Ex) or GAD2+ neurons (Eif4ebp1In), or mice mutant for Eif4ebp2 in excitatory (Eif4ebp2Ex) or inhibitory (Eif4ebp2In) neurons (corresponding to mice in Fig. 2i; 2-way ANOVA and Dunnett’s). b, In these same mice, distance travelled during the NSF task was measured. Data were expressed as average distance travelled per minute to account for differences in latency to feed (2-way ANOVA and Dunnett’s). c, Control, Eif4ebp1Ex, Eif4ebp1In, Eif4ebp2Ex and Eif4ebpIn mice were tested 6 days (144 h) after saline or Ket treatment (IP, 10 mg kg−1) in the FST (2-way ANOVA and Fisher’s LSD, n = 17, 15, 7, 10, 12, 17, 8, 9, 5, 7 from left to right). Mean ± s.e.m.; * P < 0.05 vs. control (main effect of genotype, in a) or vs. Sal-treated mice of the same genotype.
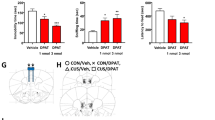
Extended Data Fig. 5 Effects of 4E-BP2 mutation in inhibitory neurons on amplitude of sEPSC and mIPSC.
a, Cumulative distribution of sEPSC amplitude in CA1 pyramidal neurons of control and Eif4ebp2In mice treated with saline or Ket (mice recorded in Fig. 3; Kolmogorov–Smirnov test). b, Amplitude of sEPSCs recorded from pyramidal neurons in CA1 of control and Eif4ebp2In mice (mice recorded in Fig. 3; 2-way ANOVA). c, Cumulative distribution of mIPSC amplitude in CA1 pyramidal neurons from mice treated as in a (mice recorded in Fig. 4; Kolmogorov–Smirnov test). d, Amplitude of mIPSC in CA1 pyramidal neurons in mice treated as in a (mice recorded in Fig. 4; 2-way ANOVA). Mean ± s.e.m.; * P < 0.05 vs. control-Sal.
Extended Data Fig. 6 The absence of 4E-BP2 in inhibitory neurons does not affect mEPSCs in pyramidal neurons.
a, Representative whole-cell recordings of mEPSCs in CA1 pyramidal neurons of mice treated with saline or Ket 24 h before (IP, 10 mg kg−1). Control and Eif4ebp2In mice were used. b, Cumulative inter-event interval (Kolmogorov–Smirnov test, n = 7, 11, 6, 17 from left from right) and c, frequency of mEPSCs measured in a (2-way ANOVA). d, Cumulative (Kolmogorov–Smirnov test) and e, average distribution of mEPSC amplitude measured in a (2-way ANOVA). Mean ± s.e.m.; * P < 0.05 vs. Eif4ebp2In-Sal.
Supplementary information
Supplementary Information
Uncropped Western blots from Extended Data Figure 1a. Uncropped images of the films for the Western blot analyses of 4E-BP1 and 4E-BP2 in the prefrontal cortex and hippocampus of wildtype (Eif4ebp1/2+/+ +/+), 4E-BP1 knock out (Eif4ebp1-/-), 4E-BP2 knock out (Eif4ebp2-/-), and 4E-BP1/2 double knock out mice (Eif4ebp1/2-/-). Images for 4E-BP1, 4E-BP2 and GAPDH are presented.
Supplementary Table 1
Details of statistical analyses.
Rights and permissions
About this article
Cite this article
Aguilar-Valles, A., De Gregorio, D., Matta-Camacho, E. et al. Antidepressant actions of ketamine engage cell-specific translation via eIF4E. Nature 590, 315–319 (2021). https://doi.org/10.1038/s41586-020-03047-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-03047-0
This article is cited by
-
Myelin-associated oligodendrocytic basic protein-dependent myelin repair confers the long-lasting antidepressant effect of ketamine
Molecular Psychiatry (2024)
-
Defective regulation of the eIF2-eIF2B translational axis underlies depressive-like behavior in mice and correlates with major depressive disorder in humans
Translational Psychiatry (2024)
-
Delta opioid receptor agonists activate PI3K–mTORC1 signaling in parvalbumin-positive interneurons in mouse infralimbic prefrontal cortex to exert acute antidepressant-lie effects
Molecular Psychiatry (2024)
-
Activity in the dorsal hippocampus-mPFC circuit modulates stress-coping strategies during inescapable stress
Experimental & Molecular Medicine (2024)
-
Ketamine Improves the Glymphatic Pathway by Reducing the Pyroptosis of Hippocampal Astrocytes in the Chronic Unpredictable Mild Stress Model
Molecular Neurobiology (2024)