Abstract
PTEN is a potent tumour suppressor, and its loss of function is frequently observed in both heritable and sporadic cancers. PTEN has phosphatase-dependent and phosphatase-independent (scaffold) activities in the cell and governs a variety of biological processes, including maintenance of genomic stability, cell survival, migration, proliferation and metabolism. Even a subtle decrease in PTEN levels and activity results in cancer susceptibility and favours tumour progression. Regulation of PTEN has therefore emerged as a subject of intense research in tumour biology. Recent discoveries, including the existence of distinct PTEN isoforms and the ability of PTEN to form dimers, have brought to light new modes of PTEN function and regulation. These milestone findings have in turn opened new therapeutic avenues for cancer prevention and treatment through restoration of PTEN tumour suppressor activity.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Li, J. et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science 275, 1943–1947 (1997).
Steck, P. A. et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat. Genet. 15, 356–362 (1997).
Li, D. M. & Sun, H. TEP1, encoded by a candidate tumor suppressor locus, is a novel protein tyrosine phosphatase regulated by transforming growth factor beta. Cancer Res. 57, 2124–2129 (1997). References 1–3 identify PTEN as the candidate tumour suppressor gene on human chromosome 10q23.
Liaw, D. et al. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat. Genet. 16, 64–67 (1997).
Marsh, D. J. et al. Germline mutations in PTEN are present in Bannayan-Zonana syndrome. Nat. Genet. 16, 333–334 (1997).
Di Cristofano, A., Pesce, B., Cordon-Cardo, C. & Pandolfi, P. P. Pten is essential for embryonic development and tumour suppression. Nat. Genet. 19, 348–355 (1998).
Suzuki, A. et al. High cancer susceptibility and embryonic lethality associated with mutation of the PTEN tumor suppressor gene in mice. Curr. Biol. 8, 1169–1178 (1998).
Podsypanina, K. et al. Mutation of Pten/Mmac1 in mice causes neoplasia in multiple organ systems. Proc. Natl Acad. Sci. USA 96, 1563–1568 (1999).
Di Cristofano, A. et al. Impaired Fas response and autoimmunity in Pten+/− mice. Science 285, 2122–2125 (1999).
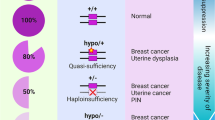
Alimonti, A. et al. Subtle variations in Pten dose determine cancer susceptibility. Nat. Genet. 42, 454–458 (2010).
Berger, A. H., Knudson, A. G. & Pandolfi, P. P. A continuum model for tumour suppression. Nature 476, 163–169 (2011). References 10 and 11 identify PTEN as a haploinsufficient and quasi-sufficient tumour suppressor gene in a continuum model for tumour suppression.
Chen, Z. et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature 436, 725–730 (2005).Reference 12 provides the first evidence that the loss of a critical tumour suppressor such as PTEN results in cellular senescence.
Galicia, V. A. et al. Expansion of hepatic tumor progenitor cells in Pten-null mice requires liver injury and is reversed by loss of AKT2. Gastroenterology 139, 2170–2182 (2010).
Song, M. S., Salmena, L. & Pandolfi, P. P. The functions and regulation of the PTEN tumour suppressor. Nat. Rev. Mol. Cell Biol. 13, 283–296 (2012). Reference 14 offers a broad overview of the functions and regulation of the PTEN tumour suppressor.
Salmena, L., Carracedo, A. & Pandolfi, P. P. Tenets of PTEN tumor suppression. Cell 133, 403–414 (2008).
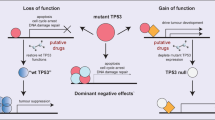
Papa, A. et al. Cancer-associated PTEN mutants act in a dominant-negative manner to suppress PTEN protein function. Cell 157, 595–610 (2014). Reference 16 demonstrates that PTEN is active in its dimer configuration at the plasma membrane.
Hopkins, B. D. et al. A secreted PTEN phosphatase that enters cells to alter signaling and survival. Science 341, 399–402 (2013).
Putz, U. et al. The tumor suppressor PTEN is exported in exosomes and has phosphatase activity in recipient cells. Sci. Signal 5, ra70 (2012). References 17 and 18 provide evidence of a function for secreted PTEN.
Keniry, M. & Parsons, R. The role of PTEN signaling perturbations in cancer and in targeted therapy. Oncogene 27, 5477–5485 (2008).
Leslie, N. R., Batty, I. H., Maccario, H., Davidson, L. & Downes, C. P. Understanding PTEN regulation: PIP2, polarity and protein stability. Oncogene 27, 5464–5476 (2008).
Lee, J. O. et al. Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell 99, 323–334 (1999).
Maehama, T. & Dixon, J. E. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 273, 13375–13378 (1998).
Stambolic, V. et al. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell 95, 29–39 (1998).
Manning, B. D. & Cantley, L. C. AKT/PKB signaling: navigating downstream. Cell 129, 1261–1274 (2007).
Liliental, J. et al. Genetic deletion of the Pten tumor suppressor gene promotes cell motility by activation of Rac1 and Cdc42 GTPases. Curr. Biol. 10, 401–404 (2000).
Funamoto, S., Meili, R., Lee, S., Parry, L. & Firtel, R. A. Spatial and temporal regulation of 3-phosphoinositides by PI3-kinase and PTEN mediates chemotaxis. Cell 109, 611–623 (2002).
Iijima, M. & Devreotes, P. Tumor suppressor PTEN mediates sensing of chemoattractant gradients. Cell 109, 599–610 (2002).
Myers, M. P. et al. P-TEN, the tumor suppressor from human chromosome 10q23, is a dual-specificity phosphatase. Proc. Natl Acad. Sci. USA 94, 9052–9057 (1997).
Gu, T. et al. CREB is a novel nuclear target of PTEN phosphatase. Cancer Res. 71, 2821–2825 (2011).
Shi, Y. et al. PTEN is a protein tyrosine phosphatase for IRS1. Nat. Struct. Mol. Biol. 21, 522–527 (2014).
Tamura, M. et al. Inhibition of cell migration, spreading, and focal adhesions by tumor suppressor PTEN. Science 280, 1614–1617 (1998).
Zhang, S. et al. Combating trastuzumab resistance by targeting SRC, a common node downstream of multiple resistance pathways. Nat. Med. 17, 461–469 (2011).
Zhang, X. C., Piccini, A., Myers, M. P., Van Aelst, L. & Tonks, N. K. Functional analysis of the protein phosphatase activity of PTEN. Biochem. J. 444, 457–464 (2012).
Davidson, L. et al. Suppression of cellular proliferation and invasion by the concerted lipid and protein phosphatase activities of PTEN. Oncogene 29, 687–697 (2010).
Myers, M. P. et al. The lipid phosphatase activity of PTEN is critical for its tumor supressor function. Proc. Natl Acad. Sci. USA 95, 13513–13518 (1998).
Blanco-Aparicio, C., Renner, O., Leal, J. F. & Carnero, A. PTEN, more than the AKT pathway. Carcinogenesis 28, 1379–1386 (2007).
Kuchay, S. et al. PTEN counteracts FBXL2 to promote IP3R3- and Ca2+-mediated apoptosis limiting tumour growth. Nature 546, 554–558 (2017).
Zhao, D. et al. Synthetic essentiality of chromatin remodelling factor CHD1 in PTEN-deficient cancer. Nature 542, 484–488 (2017). References 37 and 38 report that the functions of PTEN as a cytosolic scaffold contribute to tumour suppression.
Warburg, O. On the origin of cancer cells. Science 123, 309–314 (1956).
Vander Heiden, M. G., Cantley, L. C. & Thompson, C. B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009).
DeBerardinis, R. J. et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl Acad. Sci. USA 104, 19345–19350 (2007).
Levine, A. J. & Puzio-Kuter, A. M. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science 330, 1340–1344 (2010).
Shimobayashi, M. & Hall, M. N. Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nat. Rev. Mol. Cell Biol. 15, 155–162 (2014).
Zoncu, R., Efeyan, A. & Sabatini, D. M. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 12, 21–35 (2011).
Lizcano, J. M. & Alessi, D. R. The insulin signalling pathway. Curr. Biol. 12, R236–R238 (2002).
Miinea, C. P. et al. AS160, the Akt substrate regulating GLUT4 translocation, has a functional Rab GTPase-activating protein domain. Biochem. J. 391, 87–93 (2005).
Wong, J. T. et al. Pten (phosphatase and tensin homologue gene) haploinsufficiency promotes insulin hypersensitivity. Diabetologia 50, 395–403 (2007).
Eguez, L. et al. Full intracellular retention of GLUT4 requires AS160 Rab GTPase activating protein. Cell Metab. 2, 263–272 (2005).
Fang, M. et al. The ER UDPase ENTPD5 promotes protein N-glycosylation, the Warburg effect, and proliferation in the PTEN pathway. Cell 143, 711–724 (2010).
Puigserver, P. et al. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature 423, 550–555 (2003).
Li, X., Monks, B., Ge, Q. & Birnbaum, M. J. Akt/PKB regulates hepatic metabolism by directly inhibiting PGC-1alpha transcription coactivator. Nature 447, 1012–1016 (2007).
Duvel, K. et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol. Cell 39, 171–183 (2010).
Porstmann, T. et al. PKB/Akt induces transcription of enzymes involved in cholesterol and fatty acid biosynthesis via activation of SREBP. Oncogene 24, 6465–6481 (2005).
Porstmann, T. et al. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 8, 224–236 (2008).
Chen, M. et al. An aberrant SREBP-dependent lipogenic program promotes metastatic prostate cancer. Nat. Genet. 50, 206–218 (2018).
Horie, Y. et al. Hepatocyte-specific Pten deficiency results in steatohepatitis and hepatocellular carcinomas. J. Clin. Invest. 113, 1774–1783 (2004).
Zabala-Letona, A. et al. mTORC1-dependent AMD1 regulation sustains polyamine metabolism in prostate cancer. Nature 547, 109–113 (2017).
Mathur, D. et al. PTEN regulates glutamine flux to pyrimidine synthesis and sensitivity to dihydroorotate dehydrogenase inhibition. Cancer Discov. 7, 380–390 (2017).
Garcia-Cao, I. et al. Systemic elevation of PTEN induces a tumor-suppressive metabolic state. Cell 149, 49–62 (2012).
Ortega-Molina, A. et al. Pten positively regulates brown adipose function, energy expenditure, and longevity. Cell Metab. 15, 382–394 (2012).
Heit, B. et al. PTEN functions to ‘prioritize’ chemotactic cues and prevent ‘distraction’ in migrating neutrophils. Nat. Immunol. 9, 743–752 (2008).
Raftopoulou, M., Etienne-Manneville, S., Self, A., Nicholls, S. & Hall, A. Regulation of cell migration by the C2 domain of the tumor suppressor PTEN. Science 303, 1179–1181 (2004).
Martin-Belmonte, F. et al. PTEN-mediated apical segregation of phosphoinositides controls epithelial morphogenesis through Cdc42. Cell 128, 383–397 (2007).
Baker, S. J. PTEN enters the nuclear age. Cell 128, 25–28 (2007).
Lindsay, Y. et al. Localization of agonist-sensitive PtdIns(3,4,5)P3 reveals a nuclear pool that is insensitive to PTEN expression. J. Cell Sci. 119, 5160–5168 (2006).
Gimm, O. et al. Differential nuclear and cytoplasmic expression of PTEN in normal thyroid tissue, and benign and malignant epithelial thyroid tumors. Am. J. Pathol. 156, 1693–1700 (2000).
Shen, W. H. et al. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell 128, 157–170 (2007).
Bassi, C. et al. Nuclear PTEN controls DNA repair and sensitivity to genotoxic stress. Science 341, 395–399 (2013).
Li, A. G. et al. Mechanistic insights into maintenance of high p53 acetylation by PTEN. Mol. Cell 23, 575–587 (2006).
Song, M. S. et al. Nuclear PTEN regulates the APC-CDH1 tumor-suppressive complex in a phosphatase-independent manner. Cell 144, 187–199 (2011). References 67–70 decipher novel tumour-suppressive roles of PTEN in the nucleus, which are independent of its phosphatase activity.
Gil, A. et al. Nuclear localization of PTEN by a Ran-dependent mechanism enhances apoptosis: Involvement of an N-terminal nuclear localization domain and multiple nuclear exclusion motifs. Mol. Biol. Cell 17, 4002–4013 (2006).
Groszer, M. et al. Negative regulation of neural stem/progenitor cell proliferation by the Pten tumor suppressor gene in vivo. Science 294, 2186–2189 (2001).
Groszer, M. et al. PTEN negatively regulates neural stem cell self-renewal by modulating G0-G1 cell cycle entry. Proc. Natl Acad. Sci. USA 103, 111–116 (2006).
Wang, S. et al. Pten deletion leads to the expansion of a prostatic stem/progenitor cell subpopulation and tumor initiation. Proc. Natl Acad. Sci. USA 103, 1480–1485 (2006).
Bonaguidi, M. A. et al. In vivo clonal analysis reveals self-renewing and multipotent adult neural stem cell characteristics. Cell 145, 1142–1155 (2011).
Yilmaz, O. H. et al. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature 441, 475–482 (2006).
Zhang, J. et al. PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature 441, 518–522 (2006).
Lee, J. Y. et al. mTOR activation induces tumor suppressors that inhibit leukemogenesis and deplete hematopoietic stem cells after Pten deletion. Cell Stem Cell 7, 593–605 (2010).
Guo, W. et al. Multi-genetic events collaboratively contribute to Pten-null leukaemia stem-cell formation. Nature 453, 529–533 (2008).
Magee, J. A. et al. Temporal changes in PTEN and mTORC2 regulation of hematopoietic stem cell self-renewal and leukemia suppression. Cell Stem Cell 11, 415–428 (2012).
Kalaitzidis, D. et al. mTOR complex 1 plays critical roles in hematopoiesis and Pten-loss-evoked leukemogenesis. Cell Stem Cell 11, 429–439 (2012).
He, S. & Sharpless, N. E. Senescence in health and disease. Cell 169, 1000–1011 (2017).
Childs, B. G., Durik, M., Baker, D. J. & van Deursen, J. M. Cellular senescence in aging and age-related disease: from mechanisms to therapy. Nat. Med. 21, 1424–1435 (2015).
Campisi, J. Aging, cellular senescence, and cancer. Annu. Rev. Physiol. 75, 685–705 (2013).
Trotman, L. C. et al. Pten dose dictates cancer progression in the prostate. PLoS Biol. 1, E59 (2003).
Alimonti, A. et al. A novel type of cellular senescence that can be enhanced in mouse models and human tumor xenografts to suppress prostate tumorigenesis. J. Clin. Invest. 120, 681–693 (2010).
Nardella, C., Clohessy, J. G., Alimonti, A. & Pandolfi, P. P. Pro-senescence therapy for cancer treatment. Nat. Rev. Cancer 11, 503–511 (2011).
Di Mitri, D. et al. Tumour-infiltrating Gr-1+ myeloid cells antagonize senescence in cancer. Nature 515, 134–137 (2014).
Dorr, J. R. et al. Synthetic lethal metabolic targeting of cellular senescence in cancer therapy. Nature 501, 421–425 (2013).
Demaria, M. et al. Cellular senescence promotes adverse effects of chemotherapy and cancer relapse. Cancer Discov. 7, 165–176 (2017).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Toso, A. et al. Enhancing chemotherapy efficacy in Pten-deficient prostate tumors by activating the senescence-associated antitumor immunity. Cell Rep. 9, 75–89 (2014).
Luo, X. et al. Dual Shp2 and Pten deficiencies promote non-alcoholic steatohepatitis and genesis of liver tumor-initiating cells. Cell Rep. 17, 2979–2993 (2016).
Zhu, H. H. et al. Shp2 and Pten have antagonistic roles in myeloproliferation but cooperate to promote erythropoiesis in mammals. Proc. Natl Acad. Sci. USA 112, 13342–13347 (2015).
Trimboli, A. J. et al. Pten in stromal fibroblasts suppresses mammary epithelial tumours. Nature 461, 1084–1091 (2009).
Bronisz, A. et al. Reprogramming of the tumour microenvironment by stromal PTEN-regulated miR-320. Nat. Cell Biol. 14, 159–167 (2011).
Miething, C. et al. PTEN action in leukaemia dictated by the tissue microenvironment. Nature 510, 402–406 (2014).
Zhang, L. et al. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature 527, 100–104 (2015).
Zhou, X. P. et al. PTEN mutational spectra, expression levels, and subcellular localization in microsatellite stable and unstable colorectal cancers. Am. J. Pathol. 161, 439–447 (2002).
Mutter, G. L. et al. Altered PTEN expression as a diagnostic marker for the earliest endometrial precancers. J. Natl Cancer Inst. 92, 924–930 (2000).
Leupin, N. et al. Disparate expression of the PTEN gene: a novel finding in B-cell chronic lymphocytic leukaemia (B-CLL). Br. J. Haematol. 121, 97–100 (2003).
Tan, M. H. et al. A clinical scoring system for selection of patients for PTEN mutation testing is proposed on the basis of a prospective study of 3042 probands. Am. J. Hum. Genet. 88, 42–56 (2011).
Han, S. Y. et al. Functional evaluation of PTEN missense mutations using in vitro phosphoinositide phosphatase assay. Cancer Res. 60, 3147–3151 (2000).
Trotman, L. C. et al. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell 128, 141–156 (2007).
Salvesen, H. B. et al. PTEN methylation is associated with advanced stage and microsatellite instability in endometrial carcinoma. Int. J. Cancer 91, 22–26 (2001).
Soria, J. C. et al. Lack of PTEN expression in non-small cell lung cancer could be related to promoter methylation. Clin. Cancer Res. 8, 1178–1184 (2002).
Khan, S. et al. PTEN promoter is methylated in a proportion of invasive breast cancers. Int. J. Cancer 112, 407–410 (2004).
Mirmohammadsadegh, A. et al. Epigenetic silencing of the PTEN gene in melanoma. Cancer Res. 66, 6546–6552 (2006).
Lu, J. et al. Stem cell factor SALL4 represses the transcriptions of PTEN and SALL1 through an epigenetic repressor complex. PLoS ONE 4, e5577 (2009).
Stambolic, V. et al. Regulation of PTEN transcription by p53. Mol. Cell 8, 317–325 (2001).
Teresi, R. E., Planchon, S. M., Waite, K. A. & Eng, C. Regulation of the PTEN promoter by statins and SREBP. Hum. Mol. Genet. 17, 919–928 (2008).
Virolle, T. et al. The Egr-1 transcription factor directly activates PTEN during irradiation-induced signalling. Nat. Cell Biol. 3, 1124–1128 (2001).
Escriva, M. et al. Repression of PTEN phosphatase by Snail1 transcriptional factor during gamma radiation-induced apoptosis. Mol. Cell. Biol. 28, 1528–1540 (2008).
Lee, J. Y. et al. Id-1 activates Akt-mediated Wnt signaling and p27(Kip1) phosphorylation through PTEN inhibition. Oncogene 28, 824–831 (2009).
Yoshimi, A. et al. Evi1 represses PTEN expression and activates PI3K/AKT/mTOR via interactions with polycomb proteins. Blood 117, 3617–3628 (2011).
Xia, D. et al. Mitogen-activated protein kinase kinase-4 promotes cell survival by decreasing PTEN expression through an NF kappa B-dependent pathway. J. Biol. Chem. 282, 3507–3519 (2007).
Song, L. B. et al. The polycomb group protein Bmi-1 represses the tumor suppressor PTEN and induces epithelial-mesenchymal transition in human nasopharyngeal epithelial cells. J. Clin. Invest. 119, 3626–3636 (2009).
Hettinger, K. et al. c-Jun promotes cellular survival by suppression of PTEN. Cell Death Differ. 14, 218–229 (2007).
Whelan, J. T., Forbes, S. L. & Bertrand, F. E. CBF-1 (RBP-J kappa) binds to the PTEN promoter and regulates PTEN gene expression. Cell Cycle 6, 80–84 (2007).
Palomero, T. et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat. Med. 13, 1203–1210 (2007).
Bartel, D. P. MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 (2009).
Tay, Y., Song, S. J. & Pandolfi, P. P. The lilliputians and the giant: an emerging oncogenic microRNA network that suppresses the PTEN Tumor suppressor in vivo. Microrna 2, 127–136 (2013).
Tay, Y., Tan, S. M., Karreth, F. A., Lieberman, J. & Pandolfi, P. P. Characterization of dual PTEN and p53-targeting microRNAs identifies microRNA-638/Dnm2 as a two-hit oncogenic locus. Cell Rep 8, 714–722 (2014).
Xiao, C. et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat. Immunol. 9, 405–414 (2008).
Meng, F. et al. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 133, 647–658 (2007).
Zhang, J. G. et al. MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC). Clin. Chim. Acta 411, 846–852 (2010).
Ciuffreda, L. et al. The mitogen-activated protein kinase (MAPK) cascade controls phosphatase and tensin homolog (PTEN) expression through multiple mechanisms. J. Mol. Med. 90, 667–679 (2012).
Mu, P. et al. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 23, 2806–2811 (2009).
Chen, P. et al. MiR-200c is a cMyc-activated miRNA that promotes nasopharyngeal carcinoma by downregulating PTEN. Oncotarget 8, 5206–5218 (2017).
Poliseno, L. et al. Identification of the miR-106b~25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci. Signal 3, ra29 (2010).
Poliseno, L. et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465, 1033–1038 (2010).
Salmena, L., Poliseno, L., Tay, Y., Kats, L. & Pandolfi, P. P. A. ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146, 353–358 (2011).
Tay, Y. et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell 147, 344–357 (2011).
Karreth, F. A. et al. In vivo identification of tumor- suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell 147, 382–395 (2011). References 131–134 demonstrate the existence of a new RNA-based mechanism underlying the regulation of PTEN expression through a new RNA language whereby RNA transcripts communicate with each other through competing shared miRNAs(ceRNA model).
Du, Z. et al. Integrative analyses reveal a long noncoding RNA-mediated sponge regulatory network in prostate cancer. Nat. Commun. 7, 10982 (2016).
Li, W. et al. Downregulation of LncRNA GAS5 causes trastuzumab resistance in breast cancer. Oncotarget 7, 27778–27786 (2016).
Tay, Y. & Pandolfi, P. P. Posttranscriptional regulation of PTEN by competing endogenous RNAs. Methods Mol. Biol. 1388, 139–154 (2016).
Zarringhalam, K. et al. Identification of competing endogenous RNAs of the tumor suppressor gene PTEN: a probabilistic approach. Sci. Rep. 7, 7755 (2017).
Hershko, A. & Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 (1998).
Wickliffe, K. E., Williamson, A., Meyer, H. J., Kelly, A. & Rape, M. K11-linked ubiquitin chains as novel regulators of cell division. Trends Cell Biol. 21, 656–663 (2011).
Chen, Z. J. & Sun, L. J. Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cell 33, 275–286 (2009).
Yuan, W. C. et al. K33-linked polyubiquitination of coronin 7 by Cul3-KLHL20 ubiquitin E3 ligase regulates protein trafficking. Mol. Cell 54, 586–600 (2014).
Wang, X. et al. NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell 128, 129–139 (2007).
Amodio, N. et al. Oncogenic role of the E3 ubiquitin ligase NEDD4-1, a PTEN negative regulator, in non-small-cell lung carcinomas. Am. J. Pathol. 177, 2622–2634 (2010).
Hong, S. W. et al. p34 is a novel regulator of the oncogenic behavior of NEDD4-1 and PTEN. Cell Death Differ. 21, 146–160 (2014).
Yim, E. K. et al. Rak functions as a tumor suppressor by regulating PTEN protein stability and function. Cancer Cell 15, 304–314 (2009).
Howitt, J. et al. Ndfip1 regulates nuclear Pten import in vivo to promote neuronal survival following cerebral ischemia. J. Cell Biol. 196, 29–36 (2012).
Mund, T. & Pelham, H. R. Regulation of PTEN/Akt and MAP kinase signaling pathways by the ubiquitin ligase activators Ndfip1 and Ndfip2. Proc. Natl Acad. Sci. USA 107, 11429–11434 (2010).
Fouladkou, F. et al. The ubiquitin ligase Nedd4-1 is dispensable for the regulation of PTEN stability and localization. Proc. Natl Acad. Sci. USA 105, 8585–8590 (2008).
Maddika, S. et al. WWP2 is an E3 ubiquitin ligase for PTEN. Nat. Cell Biol. 13, 728–733 (2011).
Van Themsche, C., Leblanc, V., Parent, S. & Asselin, E. X-Linked inhibitor of apoptosis protein (XIAP) regulates PTEN ubiquitination, content, and compartmentalization. J. Biol. Chem. 284, 20462–20466 (2009).
Ahmed, S. F. et al. The chaperone-assisted E3 ligase C terminus of Hsc70-interacting protein (CHIP) targets PTEN for proteasomal degradation. J. Biol. Chem. 287, 15996–16006 (2012).
Li, N. et al. Poly-ADP ribosylation of PTEN by tankyrases promotes PTEN degradation and tumor growth. Genes Dev. 29, 157–170 (2015).
Zhang, J. et al. Deubiquitylation and stabilization of PTEN by USP13. Nat. Cell Biol. 15, 1486–1494 (2013).
Yuan, L. et al. Deubiquitylase OTUD3 regulates PTEN stability and suppresses tumorigenesis. Nat. Cell Biol. 17, 1169–1181 (2015).
Song, M. S. et al. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature 455, 813–817 (2008). References 154–156 provide evidence that PTEN is tightly regulated by a complex ubiquitylation and deubiquitylation network.
Al-Khouri, A. M., Ma, Y., Togo, S. H., Williams, S. & Mustelin, T. Cooperative phosphorylation of the tumor suppressor phosphatase and tensin homologue (PTEN) by casein kinases and glycogen synthase kinase 3beta. J. Biol. Chem. 280, 35195–35202 (2005).
Torres, J. & Pulido, R. The tumor suppressor PTEN is phosphorylated by the protein kinase CK2 at its C terminus. Implications for PTEN stability to proteasome-mediated degradation. J. Biol. Chem. 276, 993–998 (2001).
Vazquez, F., Ramaswamy, S., Nakamura, N. & Sellers, W. R. Phosphorylation of the PTEN tail regulates protein stability and function. Mol. Cell. Biol. 20, 5010–5018 (2000).
Fenton, T. R. et al. Resistance to EGF receptor inhibitors in glioblastoma mediated by phosphorylation of the PTEN tumor suppressor at tyrosine 240. Proc. Natl Acad. Sci. USA 109, 14164–14169 (2012).
Nakahata, S. et al. Loss of NDRG2 expression activates PI3K-AKT signalling via PTEN phosphorylation in ATLL and other cancers. Nat. Commun. 5, 3393 (2014).
Silva, A. et al. PTEN posttranslational inactivation and hyperactivation of the PI3K/Akt pathway sustain primary T cell leukemia viability. J. Clin. Invest. 118, 3762–3774 (2008).
Odriozola, L., Singh, G., Hoang, T. & Chan, A. M. Regulation of PTEN activity by its carboxyl-terminal autoinhibitory domain. J. Biol. Chem. 282, 23306–23315 (2007).
Rahdar, M. et al. A phosphorylation-dependent intramolecular interaction regulates the membrane association and activity of the tumor suppressor PTEN. Proc. Natl Acad. Sci. USA 106, 480–485 (2009).
Vazquez, F. et al. Phosphorylation of the PTEN tail acts as an inhibitory switch by preventing its recruitment into a protein complex. J. Biol. Chem. 276, 48627–48630 (2001).
Wu, X. et al. Evidence for regulation of the PTEN tumor suppressor by a membrane-localized multi-PDZ domain containing scaffold protein MAGI-2. Proc. Natl Acad. Sci. USA 97, 4233–4238 (2000).
Wu, Y. et al. Interaction of the tumor suppressor PTEN/MMAC with a PDZ domain of MAGI3, a novel membrane-associated guanylate kinase. J. Biol. Chem. 275, 21477–21485 (2000).
Tolkacheva, T. et al. Regulation of PTEN binding to MAGI-2 by two putative phosphorylation sites at threonine 382 and 383. Cancer Res. 61, 4985–4989 (2001).
Maccario, H., Perera, N. M., Davidson, L., Downes, C. P. & Leslie, N. R. PTEN is destabilized by phosphorylation on Thr366. Biochem. J. 405, 439–444 (2007).
Lee, S. R. et al. Reversible inactivation of the tumor suppressor PTEN by H2O2. J. Biol. Chem. 277, 20336–20342 (2002).
Cao, J. et al. Prdx1 inhibits tumorigenesis via regulating PTEN/AKT activity. EMBO J. 28, 1505–1517 (2009).
Hui, S. T. et al. Txnip balances metabolic and growth signaling via PTEN disulfide reduction. Proc. Natl Acad. Sci. USA 105, 3921–3926 (2008).
Kim, Y. C., Kitaura, H., Taira, T., Iguchi-Ariga, S. M. & Ariga, H. Oxidation of DJ-1-dependent cell transformation through direct binding of DJ-1 to PTEN. Int. J. Oncol. 35, 1331–1341 (2009).
Kwak, Y. D. et al. NO signaling and S-nitrosylation regulate PTEN inhibition in neurodegeneration. Mol. Neurodegener 5, 49 (2010).
Numajiri, N. et al. On-off system for PI3-kinase-Akt signaling through S-nitrosylation of phosphatase with sequence homology to tensin (PTEN). Proc. Natl Acad. Sci. USA 108, 10349–10354 (2011).
Gupta, A. et al. PARK2 depletion connects energy and oxidative stress to PI3K/Akt activation via PTEN S-nitrosylation. Mol. Cell 65, 999–1013 e7 (2017).
Okumura, K. et al. PCAF modulates PTEN activity. J. Biol. Chem. 281, 26562–26568 (2006).
Ikenoue, T., Inoki, K., Zhao, B. & Guan, K. L. PTEN acetylation modulates its interaction with PDZ domain. Cancer Res. 68, 6908–6912 (2008).
Chae, H. D. & Broxmeyer, H. E. SIRT1 deficiency downregulates PTEN/JNK/FOXO1 pathway to block reactive oxygen species-induced apoptosis in mouse embryonic stem cells. Stem Cells Dev. 20, 1277–1285 (2011).
Huang, J. et al. SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane. Nat. Commun. 3, 911 (2012).
Heinrich, F. et al. The PTEN tumor suppressor forms homodimers in solution. Structure 23, 1952–1957 (2015).
Masson, G. R., Perisic, O., Burke, J. E. & Williams, R. L. The intrinsically disordered tails of PTEN and PTEN-L have distinct roles in regulating substrate specificity and membrane activity. Biochem. J. 473, 135–144 (2016).
Fine, B. et al. Activation of the PI3K pathway in cancer through inhibition of PTEN by exchange factor P-REX2a. Science 325, 1261–1265 (2009).
He, L., Ingram, A., Rybak, A. P. & Tang, D. Shank-interacting protein-like 1 promotes tumorigenesis via PTEN inhibition in human tumor cells. J. Clin. Invest. 120, 2094–2108 (2010).
He, L. et al. alpha-Mannosidase 2C1 attenuates PTEN function in prostate cancer cells. Nat. Commun. 2, 307 (2011).
Cao, J. et al. MC1R is a potent regulator of PTEN after UV exposure in melanocytes. Mol. Cell 51, 409–422 (2013).
Okahara, F. et al. Critical role of PICT-1, a tumor suppressor candidate, in phosphatidylinositol 3,4,5-trisphosphate signals and tumorigenic transformation. Mol. Biol. Cell 17, 4888–4895 (2006).
Sheng, Z. M. et al. Multiple regions of chromosome 6q affected by loss of heterozygosity in primary human breast carcinomas. Br. J. Cancer 73, 144–147 (1996).
Okahara, F., Ikawa, H., Kanaho, Y. & Maehama, T. Regulation of PTEN phosphorylation and stability by a tumor suppressor candidate protein. J. Biol. Chem. 279, 45300–45303 (2004).
Cotter, L. et al. Dlg1-PTEN interaction regulates myelin thickness to prevent damaging peripheral nerve overmyelination. Science 328, 1415–1418 (2010).
Sandoval, G. J. et al. Novel mechanism of tumor suppression by polarity gene discs large 1 (DLG1) revealed in a murine model of pediatric B-ALL. Cancer Immunol. Res. 1, 426–437 (2013).
Li, Z. et al. Regulation of PTEN by Rho small GTPases. Nat. Cell Biol. 7, 399–404 (2005).
Lima-Fernandes, E. et al. Distinct functional outputs of PTEN signalling are controlled by dynamic association with beta-arrestins. EMBO J 30, 2557–2568 (2011).
van Diepen, M. T. et al. MyosinV controls PTEN function and neuronal cell size. Nat. Cell Biol. 11, 1191–1196 (2009).
Liang, H. et al. PTENalpha, a PTEN isoform translated through alternative initiation, regulates mitochondrial function and energy metabolism. Cell Metab. 19, 836–848 (2014).
Liang, H. et al. PTENbeta is an alternatively translated isoform of PTEN that regulates rDNA transcription. Nat. Commun. 8, 14771 (2017).
Chen, M. et al. Deregulated PP1alpha phosphatase activity towards MAPK activation is antagonized by a tumor suppressive failsafe mechanism. Nat. Commun. 9, 159 (2018).
Altinoglu, S. A., Wang, M., Li, K. Q., Li, Y. & Xu, Q. Intracellular delivery of the PTEN protein using cationic lipidoids for cancer therapy. Biomater. Sci. 4, 1773–1780 (2016).
Nguyen, H. N. et al. Engineering ePTEN, an enhanced PTEN with increased tumor suppressor activities. Proc. Natl Acad. Sci. USA 111, E2684–E2693 (2014).
Acknowledgements
The authors thank all the members of the Pandolfi laboratory for their constructive comments and L. Southwood and E. Stack for editing the manuscript. This work was supported by funding from the National Cancer Institute (R35CA197529 to P.P.P.) and the Department of Defense PCRP Postdoctoral Training award to Y.-R.L. and M.C.
Reviewer information
Nature Reviews Molecular Cell Biology thanks G.-S. Feng and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
Y.-R.L., M.C. and P.P.P. researched data for the article, contributed to discussion of the content, wrote the article and reviewed and/or edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related Links
PhosphoSitePlus: https://www.phosphosite.org/homeAction;.Sanger
Institute Catalogue of Somatic Mutations in Cancer (COSMIC): http://cancer.sanger.ac.uk/cosmic/search?q=pten
Glossary
- PTEN hamartoma tumour syndromes
-
(PHTS). Encompass a spectrum of disorders caused by germline mutations of the PTEN gene. These disorders are characterized by multiple hamartomas that can affect various organs. Hamartoma is a general term for a benign tumour-like malformation composed of mature cells in a tissue that has grown in a disorganized manner as a result of developmental defects.
- Haploinsufficient
-
A situation in a diploid organism when one copy of a gene is inactivated due to mutation or deletion but the product of the remaining normal allele is not sufficient for normal cellular functional output (for example, suppression of tumorigenesis), resulting in an abnormal phenotype or diseased state (for example, tumour initiation).
- PI3K–AKT–mTOR pathway
-
A central signalling pathway that integrates both extracellular and intracellular signals to control cellular metabolism, growth, proliferation, cancer and longevity. Its activation contributes to the pathogenesis of many tumour types. This pathway is antagonized by various factors and, notably, by PTEN.
- Small-angle X-ray scattering (SAXS) analysis
-
A powerful technique used to study protein structure and interactions that is capable of delivering structural information in dimensions between 1 and 100 nm.
- PEST (Pro, Glu, Ser, Thr) sequences
-
A peptide sequence that is rich in proline (P), glutamic acid (E), serine (S) and threonine (T). Proteins possessing this sequence display a short intracellular half-life. Therefore, it is hypothesized that this sequence acts as a signal peptide for protein degradation.
- PDZ domain
-
A common structural domain of 80–90 amino acids that serves as a protein–protein interaction motif and is found in signalling proteins in bacteria, yeast, plants, viruses and animals.
- Dual-specificity protein phosphatase
-
A phosphatase that can act on both tyrosine and serine/threonine residues.
- Annexin 2
-
A Ca2+-regulated membrane protein and an F-actin-binding protein enriched at actin assembly sites on the plasma membrane and on endosomal vesicles. It is involved in diverse cellular functions, including cell motility and endocytosis.
- Major vault protein
-
(MVP). Large multi-subunit ribonucleoprotein particle, which is considered to be a general carrier for nucleocytoplasmic transport.
- RAN-GTPase
-
A GTP-binding protein involved in nucleocytoplasmic transport. It is required for the import of proteins into the nucleus and for RNA export.
- Centromere
-
The part of a chromosome that links sister chromatids. The physical role of the centromere is to act as the site of assembly of the kinetochores — a highly complex multiprotein structure that is responsible for chromosome segregation.
- Anaphase-promoting complex
-
(APC/C). Also known as the cyclosome. An E3 ubiquitin ligase that has a crucial function in the regulation of the mitotic cell cycle through targeting key mitotic regulators for degradation by the 26 S proteasome.
- Dentate gyrus
-
A part of the hippocampus that is thought to contribute to the formation of new episodic memories, the spontaneous exploration of unknown environments and other functions.
- β-catenin
-
A subunit of the cadherin protein complex that constitutes adherens junctions and a key downstream effector in the canonical WNT signalling pathway.
- ETS2
-
A transcription factor that binds specifically to the DNA GGAA/T core motif in gene promoters and stimulates transcription of genes involved in development and apoptosis.
- Senescence-associated secretory phenotype
-
(SASP). Effects mediated by the secretion of a range of proteins, including cytokines, chemokines and proteases by senescent cells. These effects are extremely diverse and include both autocrine and paracrine signalling, pro-tumorigenic and tumour-suppressive effects and pro-inflammatory and anti-inflammatory signalling.
- Exosomes
-
Cell-derived vesicles that are present and secreted out of mammalian cells.
- Competitive endogenous RNA
-
(ceRNA). A type of RNA that communicates with and regulates other RNA transcripts by competing for shared microRNAs.
- Sporadic cancers
-
Cancers that occur in individuals who do not have a family history of that cancer or an inherited change in their DNA that would increase their risk of that cancer.
- Cowden disease
-
An autosomal dominant multiple hamartoma syndrome that results most commonly from a mutation in the PTEN gene. The disease was named after the family in which it was first reported. Although the tumours are largely benign, individuals with Cowden syndrome have an increased risk of developing several types of cancer, including cancers of the breast, thyroid and uterus.
- ERK
-
A serine/threonine-specific protein kinase that is involved in directing cellular responses to a diverse array of stimuli, such as mitogens, osmotic stress, heat shock and pro-inflammatory cytokines. It regulates cellular functions, including proliferation, gene expression, differentiation, mitosis and cell survival.
- Pseudogene
-
A genetic unit transcribed into non-coding RNA that has a counterpart in another gene from which it is derived. The expressed pseudogenes have promoters, CpG islands and splice sites, although some do not contain introns because they were generated from mRNA retrotransposition. Pseudogenes have often lost their ability to encode proteins, for which they are often referred to as ‘junk DNA’. They have now been functionally resurrected and attributed to a number of non-coding RNA functions, including their ability to act as competitive endogenous RNAs.
- Nucleophile
-
A chemical species that can donate an electron pair to an electrophile to form a chemical bond in relation to a reaction.
- Anchorage-independent cell growth
-
A type of growth where cells grow in the absence of a stable surface to which they can adhere. It is a key aspect of the neoplastic transformation, particularly with respect to metastatic potential.
- Schwann cells
-
Cells in the peripheral nervous system that produce the myelin sheath around neuronal axons and function to support neurons. They are named after German physiologist Theodor Schwann, who discovered them in the 19th century.
- Imaginal discs
-
Sac-like epithelial structures that are found inside the larvae of insects and become a portion of the outside of the adult insect after pupal transformation.
- Sciatic nerve
-
The largest single nerve in humans and animals. It runs from each side of the lower spine through deep in the buttock into the back of the thigh and all the way down to the foot. It serves a vital role in connecting the spinal cord with the leg and foot muscles.
- RHOA
-
A member of the RHO family of small GTPases, which function as molecular switches in signal transduction cascades. The RHOA protein promotes reorganization of the actin cytoskeleton and regulates cell shape, attachment and motility and is associated with cancer cell proliferation and metastasis.
- β-arrestins
-
Multifunctional adaptor proteins that are best known for their ability to desensitize G protein-coupled receptors, thereby regulating a diverse array of cellular functions downstream of G protein-coupled receptors.
- PTEN-induced putative kinase 1
-
(PINK1). A mitochondrial serine/threonine-protein kinase encoded by the PINK1 gene. PINK1 activity triggers the binding of the parkin protein to depolarized mitochondria, in turn inducing mitophagy. Mutations in this gene result in autosomal recessive Parkinson disease.
- Nucleolin
-
The major nucleolar protein of growing eukaryotic cells. This protein is associated with chromatin and pre-ribosomal particles and functions to regulate ribosomal RNA transcription and ribosome assembly.
- Lipidoids
-
Any materials having characteristics of a lipid.
Rights and permissions
About this article
Cite this article
Lee, YR., Chen, M. & Pandolfi, P.P. The functions and regulation of the PTEN tumour suppressor: new modes and prospects. Nat Rev Mol Cell Biol 19, 547–562 (2018). https://doi.org/10.1038/s41580-018-0015-0
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41580-018-0015-0
This article is cited by
-
Phenotypic rescue via mTOR inhibition in neuron-specific Pten knockout mice reveals AKT and mTORC1-site specific changes
Molecular Psychiatry (2025)
-
Advances in cancer genomics and precision oncology
Genes & Genomics (2025)
-
Inhibition of hemangioma development by regulating the VEGF/VEGFR autocrine loop via the miR-494/PTEN pathway
Discover Oncology (2025)
-
WWP1 inhibition suppresses the proliferation of pancreatic cancer cells by regulating the PI3K-AKT pathway
Journal of Gastroenterology (2025)
-
Tumorigenesis of basal muscle invasive bladder cancer was mediated by PTEN protein degradation resulting from SNHG1 upregulation
Journal of Experimental & Clinical Cancer Research (2024)