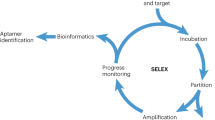
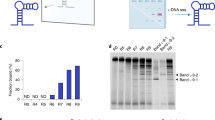
Abstract
Aptamers are nucleic acid molecules that mimic antibodies by folding into complex 3D shapes that bind to specific targets. Although some aptamers exist naturally as the ligand-binding elements of riboswitches, most are generated in vitro and can be tailored for a specific target. Relative to antibodies, aptamers benefit from their ease of generation, low production cost, low batch-to-batch variability, reversible folding properties and low immunogenicity. However, the true value of aptamers lies in the simplicity by which these molecules can be engineered into sensors, actuators and other devices that are often central to emerging technologies. This Review examines changing trends in aptamer technology by analysing the first quarter century of aptamer data that is available in the scientific literature (1990–2015). We highlight specific examples that showcase the use of aptamers in key applications, discuss challenges that have impeded the success of aptamers in practical applications, provide suggestions for choosing chemical modifications that can lead to enhanced activity or stability, and propose standards for the characterization of aptamers in the scientific literature.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Venter, J. C. et al. The sequence of the human genome. Science 291, 1304–1351 (2001).
Lander, E. S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Doudna, J. A. & Cech, T. R. The chemical repertoire of natural ribozymes. Nature 418, 222–228 (2002).
Sonenberg, N. & Hinnebusch, A. G. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136, 731–745 (2009).
Zovoilis, A., Cifuentes-Rojas, C., Chu, H. P., Hernandez, A. J. & Lee, J. T. Destabilization of B2 RNA by EZH2 activates the stress response. Cell 167, 1788–1802 (2016).
Moore, P. B. & Steitz, T. A. The involvement of RNA in ribosome function. Nature 418, 229–235 (2002).
Onoa, B. & Tinoco, I. Jr. RNA folding and unfolding. Curr. Opin. Struct. Biol. 14, 374–379 (2004).
Caruthers, M. H. Gene synthesis machines: DNA chemistry and its uses. Science 230, 281–285 (1985).
Saiki, R. et al. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239, 487–491 (1988).
Wilson, D. S. & Szostak, J. W. In vitro selection of functional nucleic acids. Annu. Rev. Biochem. 68, 611–647 (1999).
Ellington, A. D. & Szostak, J. W. In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822 (1990).
Tuerk, C. & Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510 (1990).
Robertson, D. L. & Joyce, G. F. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature 344, 467–468 (1990).
Levine, H. A. & Nilsen-Hamilton, M. A mathematical analysis of SELEX. Comput. Biol. Chem. 31, 11–35 (2007).
Szostak, J. W. In vitro genetics. Trends Biochem. Sci. 17, 89–93 (1992).
Joyce, G. F. In vitro evolution of nucleic acids. Curr. Opin. Struct. Biol. 4, 331–336 (1994).
Jayasena, S. D. Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 45, 1628–1650 (1999).
Ruigrok, V. J., Levisson, M., Eppink, M. H. M., Smidt, H. & van der Oost, J. Alternative affinity tools: more attractive than antibodies. Biochem. J. 436, 1–13 (2011).
Binz, H. K. et al. High-affinity binders selected from designed ankyrin repeat protein libraries. Nat. Biotechnol. 22, 575–582 (2004).
Hey, T., Fiedler, E., Rudolph, R. & Fiedler, M. Artificial, non-antibody binding proteins for pharmaceutical and industrial applications. Trends Biotechnol. 23, 514–522 (2005).
Hoogenboom, H. R. Selecting and screening recombinant antibody libraries. Nat. Biotechnol. 23, 1105–1116 (2005).
Sidhu, S. S. & Fellouse, F. A. Synthetic therapeutic antibodies. Nat. Chem. Biol. 2, 682–688 (2006).
Carothers, J. M., Goler, J. A., Juminaga, D. & Keasling, J. D. Model-driven engineering of RNA devices to quantitatively program gene expression. Science 334, 1716–1719 (2011).
Marx, V. Finding the right antibody for the job. Nat. Methods 10, 703–707 (2013).
Keefe, A. D., Pai, S. & Ellington, A. D. Aptamers as therapeutics. Nat. Rev. Drug Discov. 9, 537–550 (2010).
Nimjee, S. M., Rusconi, C. P. & Sullenger, B. A. Aptamers: an emerging class of therapeutics. Annu. Rev. Med. 56, 555–583 (2005).
Zhou, J. & Rossi, J. Aptamers as targeted therapeutics: current potential and challenges. Nat. Rev. Drug Discov. 16, 181–202 (2017).
Rusconi, C. P. et al. Antidote-mediated control of an anticoagulant aptamer in vivo. Nat. Biotechnol 22, 1423–1428 (2004).
Tan, W., Donovan, M. J. & Jiang, J. Aptamers from cell-based selection for bioanalytical applications. Chem. Rev. 113, 2842–2862 (2013).
Sismour, A. M. et al. PCR amplification of DNA containing non-standard base pairs by variants of reverse transcriptase from Human Immunodeficiency Virus-1. Nucleic Acids Res. 32, 728–735 (2004).
Pinheiro, A. V., Han, D., Shih, W. M. & Yan, H. Challenges and opportunities for structural DNA nanotechnology. Nat. Nanotechnol. 6, 763–772 (2011).
Lund, K. et al. Molecular robots guided by prescriptive landscapes. Nature 465, 206–210 (2010).
Mayer, G. The chemical biology of aptamers. Angew. Chem. Int. Ed. 48, 2672–2689 (2009).
Cho, E. J., Lee, J.-W. & Ellington, A. D. Applications of aptamers as sensors. Annu. Rev. Anal. Chem. 2, 241–264 (2009).
Ozer, A., Pagano, J. M. & Lis, J. T. New technologies provide quantum changes in the scale, speed, and success of SELEX methods and aptamer characterization. Mol. Ther. Nucleic Acids 3, e183 (2014).
McKeague, M. et al. Analysis of in vitro aptamer selection parameters. J. Mol. Evol. 81, 150–161 (2015).
Sassanfar, M. & Szostak, J. W. An RNA motif that binds ATP. Nature 364, 550–553 (1993).
Wilson, D. S., Keefe, A. D. & Szostak, J. W. The use of mRNA display to select high-affinity protein-binding peptides. Proc. Natl Acad. Sci. USA 98, 3750–3755 (2001).
Vaish, N. K., Larralde, R., Fraley, A. W., Szostak, J. W. & McLaughlin, L. W. A novel, modification-dependent ATP-binding aptamer selected from an RNA library incorporating a cationic functionality. Biochemistry 42, 8842–8851 (2003).
Eaton, B. E. et al. Post-SELEX combinatorial optimization of aptamers. Bioorg. Med. Chem. 5, 1087–1096 (1997).
Khvorova, A. & Watts, J. K. The chemical evolution of oligonucleotide therapies of clinical utility. Nat. Biotechnol. 35, 238–248 (2017).
Ng, E. W. M. et al. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 5, 123–132 (2006).
Ruckman, J. et al. 2′-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J. Biol. Chem. 273, 20556–20567 (1998).
Group, C. R. et al. Ranibizumab and bevacizumab for neovascular age-related macular degeneration. N. Engl. J. Med. 364, 1897–1908 (2011).
Heier, J. S. et al. Intravitreal aflibercept (VEGF trap-eye) in wet age-related macular degeneration. Ophthalmology 119, 2537–2548 (2012).
Abeydeera, N. D. et al. Evoking picomolar binding in RNA by a single phosphorodithioate linkage. Nucleic Acids Res. 44, 8052–8064 (2016).
Williams, B. A. R. et al. Creating protein affinity reagents by combining peptide ligands on synthetic DNA scaffolds. J. Am. Chem. Soc. 131, 17233–17241 (2009).
Bittker, J. A., Phillips, K. J. & Liu, D. R. Recent advances in the in vitro evolution of nucleic acids. Curr. Opin. Chem. Biol. 6, 367–374 (2002).
Yang, K. A. et al. Recognition and sensing of low-epitope targets via ternary complexes with oligonucleotides and synthetic receptors. Nat. Chem. 6, 1003–1008 (2014).
Ferguson, B. S. et al. Real-time, aptamer-based tracking of circulating therapeutic agents in living animals. Sci. Transl Med. 5, 213ra165 (2013).
Amaya-Gonzalez, S., de-los-Santos-Alvarez, N., Miranda-Ordieres, A. J. & Lobo-Castanon, M. J. Aptamer-based analysis: a promising alternative for food safety control. Sensors 13, 16292–16311 (2013).
Hayat, A. & Marty, J. L. Aptamer based electrochemical sensors for emerging environmental pollutants. Front. Chem. 2, 41 (2014).
Smiley, D. A. & Becker, R. C. Factor IXa as a target for anticoagulation in thrombotic disorders and conditions. Drug Discov. Today 19, 1445–1453 (2014).
Rusconi, C. P. et al. RNA aptamers as reversible antagonists of coagulation factor IXa. Nature 419, 90–94 (2002).
Povsic, T. J. et al. Use of the REG1 anticoagulation system in patients with acute coronary syndromes undergoing percutaneous coronary intervention: results from the phase II RADAR-PCI study. EuroIntervention 10, 431–438 (2014).
Povsic, T. J. et al. A phase 2, randomized, partially blinded, active-controlled study assessing the efficacy and safety of variable anticoagulation reversal using the REG1 system in patients with acute coronary syndromes: results of the RADAR trial. Eur. Heart J. 34, 2481–2489 (2013).
Hoellenriegel, J. et al. The Spiegelmer NOX-A12, a novel CXCL12 inhibitor, interferes with chronic lymphocytic leukemia cell motility and causes chemosensitization. Blood 123, 1032–1039 (2014).
Marasca, R. & Maffei, R. NOX-A12: mobilizing CLL away from home. Blood 123, 952–953 (2014).
Vater, A. & Klussmann, S. Turning mirror-image oligonucleotides into drugs: the evolution of Spiegelmer therapeutics. Drug Discov. Today 20, 147–155 (2015).
Klussmann, S., Nolte, A., Bald, R., Erdmann, V. A. & Furste, J. P. Mirror-image RNA that binds D-adenosine. Nat. Biotechnol. 14, 1112–1115 (1996).
Sczepanski, J. T. & Joyce, G. F. Specific inhibition of microRNA processing using L-RNA aptamers. J. Am. Chem. Soc. 137, 16032–16037 (2015).
Kruspe, S., Mittelberger, F., Szameit, K. & Hahn, U. Aptamers as drug delivery vehicles. ChemMedChem 9, 1998–2011 (2014).
Olson, W. C., Heston, W. D. & Rajasekaran, A. K. Clinical trials of cancer therapies targeting prostate-specific membrane antigen. Rev. Recent Clin. Trials 2, 182–190 (2007).
Lupold, S. E., Hicke, B. J., Lin, Y. & Coffey, D. S. Identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigen. Cancer Res. 62, 4029–4033 (2002).
Pastor, F., Kolonias, D., Giangrande, P. H. & Gilboa, E. Induction of tumour immunity by targeted inhibition of nonsense-mediated mRNA decay. Nature 465, 227–230 (2010).
Liang, C. et al. Aptamer-functionalized lipid nanoparticles targeting osteoblasts as a novel RNA interference-based bone anabolic strategy. Nat. Med. 21, 288–294 (2015).
Douek, D. C., Roederer, M. & Koup, R. A. Emerging concepts in the immunopathogenesis of AIDS. Annu. Rev. Med. 60, 471–484 (2009).
Tang, J. & Kaslow, R. A. The impact of host genetics on HIV infection and disease progression in the era of highly active antiretroviral therapy. AIDS 17 (Suppl. 4), 51–60 (2003).
Zhou, J. et al. Selection, characterization and application of new RNA HIV gp 120 aptamers for facile delivery of Dicer substrate siRNAs into HIV infected cells. Nucleic Acids Res. 37, 3094–3109 (2009).
Zhou, J., Li, H., Li, S., Zaia, J. & Rossi, J. J. Novel dual inhibitory function aptamer–siRNA delivery system for HIV-1 therapy. Mol. Ther. 16, 1481–1489 (2008).
Zhou, J. et al. Functional in vivo delivery of multiplexed anti-HIV-1 siRNAs via a chemically synthesized aptamer with a sticky bridge. Mol. Ther 21, 192–200 (2013).
Kahsai, A. W. et al. Conformationally selective RNA aptamers allosterically modulate the β2-adrenoceptor. Nat. Chem. Biol. 12, 709–716 (2016).
Lefkowitz, R. J. A brief history of G-protein coupled receptors (Nobel Lecture). Angew. Chem. Int. Ed. 52, 6366–6378 (2013).
Koehn, F. E. & Carter, G. T. The evolving role of natural products in drug discovery. Nat. Rev. Drug Discov. 4, 206–220 (2005).
Bastian, A. A., Marcozzi, A. & Herrmann, A. Selective transformations of complex molecules are enabled by aptameric protective groups. Nat. Chem. 4, 789–793 (2012).
Sefah, K. et al. In vitro selection with artificial expanded genetic information systems. Proc. Natl Acad. Sci. USA 111, 1449–1454 (2014).
Kimoto, M., Yamashige, R., Matsunaga, K. I., Yokoyama, S. & Hirao, I. Generation of high-affinity DNA aptamers using an expanded genetic alphabet. Nat. Biotechnol. 31, 453–457 (2013).
Matsunaga, K. I., Kimoto, M. & Hirao, I. High-affinity DNA aptamer generation targeting von Willebrand factor A1-domain by genetic alphabet expansion for systematic evolution of ligands by exponential enrichment using two types of libraries composed of five different bases. J. Am. Chem. Soc. 139, 324–334 (2017).
Tolle, F., Brändle, G. M., Natzner, D. & Mayer, G. A versatile approach towards nucleobase-modified aptamers. Angew. Chem. Int. Ed. 54, 10971–10974 (2015).
Shui, B. et al. RNA aptamers that functionally interact with green fluorescent protein and its derivatives. Nucleic Acids Res. 40, e39 (2012).
Gold, L. et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One 5, e15004 (2010).
Vaught, J. D. et al. Expanding the chemistry of DNA for in vitro selection. J. Am. Chem. Soc. 132, 4141–4151 (2010).
Gupta, S. et al. Chemically modified DNA aptamers bind interleukin-6 with high affinity and inhibit signaling by blocking its interaction with interleukin-6 receptor. J. Biol. Chem. 289, 8706–8719 (2014).
Gelinas, A. D., Davies, D. R. & Janjic, N. Embracing proteins: structural themes in aptamer–protein complexes. Curr. Opin. Struct. Biol. 36, 122–132 (2016).
Gawande, B. N. et al. Selection of DNA aptamers with two modified bases. Proc. Natl Acad. Sci. USA 114, 2898–2903 (2017).
Ostroff, R. M. et al. Unlocking biomarker discovery: large scale application of aptamer proteomic technology for early detection of lung cancer. PLoS One 5, e15003 (2010).
Hili, R., Niu, J. & Liu, D. R. DNA ligase-mediated translation of DNA into densely functionalized nucleic acid polymers. J. Am. Chem. Soc. 135, 98–101 (2013).
Guo, C., Watkins, C. P. & Hili, R. Sequence-defined scaffolding of peptides on nucleic acid polymers. J. Am. Chem. Soc. 137, 11191–11196 (2015).
Kong, D., Lei, Y., Yeung, W. & Hili, R. Enzymatic synthesis of sequence-defined synthetic nucleic acid polymers with diverse functional groups. Angew. Chem. Int. Ed. 55, 13164–13168 (2016).
Zhao, H. & Arnold, F. H. Combinatorial protein design: strategies for screening protein libraries. Curr. Opin. Struct. Biol. 7, 480–485 (1997).
Bordeaux, J. et al. Antibody validation. Biotechniques 48, 197–209 (2010).
Marx, V. Calling the next generation of affinity reagents. Nat. Methods 10, 829–833 (2013).
Mi, J. et al. In vivo selection of tumor-targeting RNA motifs. Nat. Chem. Biol. 6, 22–24 (2010).
Cheng, C., Chen, Y. H., Lennox, K. A., Behlke, M. A. & Davidson, B. L. In vivo SELEX for identification of brain-penetrating aptamers. Mol. Ther. Nucleic Acids 2, e67 (2013).
Wang, J. et al. Multiparameter particle display (MPPD): a quantitative screening method for the discovery of highly specific aptamers. Angew. Chem. Int. Ed. 56, 744–747 (2017).
Wang, J. et al. Particle display: a quantitative screening method for generating high-affinity aptamers. Angew. Chem. Int. Ed. 126, 4896–4901 (2014).
Griffin, L. C., Tidmarsh, G. F., Bock, L. C., Toole, J. J. & Leung, L. L. In vivo anticoagulant properties of a novel nucleotide-based thrombin inhibitor and demonstration of regional anticoagulation in extracorporeal circuits. Blood 81, 3271–3276 (1993).
Keefe, A. D. & Cload, S. T. SELEX with modified nucleotides. Curr. Opin. Chem. Biol. 12, 448–456 (2008).
Lin, Y., Qiu, Q., Gill, S. C. & Jayasena, S. D. Modified RNA sequence pools for in vitro selection. Nucleic Acids Res. 22, 5229–5234 (1994).
Burmeister, P. E. et al. Direct in vitro selection of a 2′-O-methyl aptamer to VEGF. Chem. Biol. 12, 25–33 (2005).
Burmeister, P. E. et al. 2′-Deoxy purine, 2′-O-methyl pyrimidine (dRmY) aptamers as candidate therapeutics. Oligonucleotides 16, 337–351 (2006).
Thirunavukarasu, D., Chen, T., Liu, Z., Hongdilokkul, N. & Romesberg, F. E. Selection of 2′-fluoro-modified aptamers with optimized properties. J. Am. Chem. Soc. 139, 2892–2895 (2017).
Cummins, L. L. et al. Characterization of fully 2′-modified oligoribonucleotide hetero- and homoduplex hybridization and nuclease sensitivity. Nucleic Acids Res. 23, 2019–2024 (1995).
Noronha, A. M. et al. Synthesis and biophysical properties of arabinonucleic acids (ANA): circular dichroic spectra, melting temperatures, and ribonuclease H susceptibility of ANA•RNA hybrid duplexes. Biochemistry 39, 7050–7062 (2000).
Joyce, G. F. Toward an alternative biology. Science 336, 307–308 (2012).
Legrain, P. et al. The human proteome project: current state and future direction. Mol. Cell. Proteomics 10, M111.009993 (2011).
Wang, Z., Xu, W., Liu, L. & Zhu, T. F. A synthetic molecular system capable of mirror-image genetic replication and transcription. Nat. Chem. 8, 698–704 (2016).
Pech, A. et al. A thermostable D-polymerase for mirror-image PCR. Nucleic Acids Res. 45, 3997–4005 (2017).
Larsen, A. C. et al. A general strategy for expanding polymerase function by droplet microfluidics. Nat. Commun. 7, 11235 (2016).
Ghadessy, F. J., Ong, J. L. & Holliger, P. Directed evolution of polymerase function by compartmentalized self-replication. Proc. Natl Acad. Sci. USA 98, 4552–4557 (2001).
Houlihan, G., Arangundy-Franklin, S. & Holliger, P. Exploring the chemistry of genetic information storage and propagation through polymerase engineering. Acc. Chem. Res. 50, 1079–1087 (2017).
Chen, T. & Romesberg, F. E. Directed polymerase evolution. FEBS Lett. 588, 219–229 (2014).
Taylor, A. I. & Holliger, P. Directed evolution of artificial enzymes (XNAzymes) from diverse repertoires of synthetic genetic polymers. Nat. Protoc. 10, 1625–1642 (2015).
Pinheiro, V. B. et al. Synthetic genetic polymers capable of heredity and evolution. Science 336, 341–344 (2012).
Alves Ferreira-Bravo, I., Cozens, C., Holliger, P. & DeStefano, J. J. Selection of 2′-deoxy-2′-fluoroarabinonucleotide (FANA) aptamers that bind HIV-1 reverse transcriptase with picomolar affinity. Nucleic Acids Res. 43, 9587–9599 (2015).
Yu, H., Zhang, S. & Chaput, J. C. Darwinian evolution of an alternative genetic system provides support for TNA as an RNA progenitor. Nat. Chem. 4, 183–187 (2012).
Dunn, M. R. & Chaput, J. C. Reverse transcription of threose nucleic acid by a naturally occurring DNA polymerase. ChemBioChem 17, 1804–1808 (2016).
Schöning, K. U. et al. Chemical etiology of nucleic acid structure: the α-threofuranosyl-(3′→2′) oligonucleotide system. Science 290, 1347–1351 (2000).
Ebert, M. O., Mang, C., Krishnamurthy, R., Eschenmoser, A. & Jaun, B. The structure of a TNA–TNA complex in solution: NMR study of the octamer duplex derived from α-(L)-threofuranosyl-(3′-2′)-CGAATTCG. J. Am. Chem. Soc. 130, 15105–15115 (2008).
Culbertson, M. C. et al. Evaluating TNA stability under simulated physiological conditions. Bioorg. Med. Chem. Lett. 26, 2418–2421 (2016).
Tizei, P. A., Csibra, E., Torres, L. & Pinheiro, V. B. Selection platforms for directed evolution in synthetic biology. Biochem. Soc. Trans. 44, 1165–1175 (2016).
Mei, H. et al. Synthesis and polymerase activity of a fluorescent cytidine TNA triphosphate analogue. Nucleic Acids Res. 45, 5629–5638 (2017).
Mendonsa, S. D. & Bowser, M. T. In vitro evolution of functional DNA using capillary electrophoresis. J. Am. Chem. Soc. 126, 20–21 (2004).
Ouellet, E., Foley, J. H., Conway, E. M. & Haynes, C. Hi-Fi SELEX: a high-fidelity digital-PCR based therapeutic aptamer discovery platform. Biotechnol. Bioeng. 112, 1506–1522 (2015).
Williams, R. et al. Amplification of complex gene libraries by emulsion PCR. Nat. Methods 3, 545–550 (2006).
Cox, J. C. et al. Automated selection of aptamers against protein targets translated in vitro: from gene to aptamer. Nucleic Acids Res. 30, e108 (2002).
Bradbury, A. & Pluckthun, A. Reproducibility: standardize antibodies used in research. Nature 518, 27–29 (2015).
Barrett, S. E. et al. An in vivo evaluation of amphiphilic, biodegradable peptide copolymers as siRNA delivery agents. Int. J. Pharm. 466, 58–67 (2014).
Long, S. B., Long, M. B., White, R. R. & Sullenger, B. A. Crystal structure of an RNA aptamer bound to thrombin. RNA 14, 2504–2512 (2008).
Acknowledgements
The authors thank members of the Chaput laboratory for helpful comments and suggestions, and N. Chim for assistance with the figures. This work was supported by the Defense Advanced Research Projects Agency (DARPA) Folded Non-Natural Polymers with Biological Function (Fold Fx) Program (Award No.N66001-16-2-4061).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Supplementary information S1–S21 (tables)
Raw and normalized data for aptamer trends presented in Figure 1b. (XLSX 335 kb)
Glossary
- Phenotypes
-
Observable characteristics, such as binding or catalytic activity, that are encoded in the sequence of a nucleic acid or protein.
- Genotypes
-
Genetic sequences that encode phenotypes.
- Antibodies
-
Protein affinity reagents produced by the immune system that can be made to recognize a wide range of targets called antigens.
- Aptagenic
-
A target that is known to produce an aptamer by in vitro selection.
- Dissociation constant
-
The equilibrium constant for the dissociation of the target– aptamer complex (Kd = [target] [aptamer]/[target–aptamer]).
- Lysate
-
The contents of a cell produced by cell lysis.
- cDNA
-
The complementary DNA sequence that results when RNA (or xeno-nucleic acid (XNA)) is reverse transcribed into DNA.
- Doped library
-
A nucleic acid library that contains variants of a single sequence, which feature a small fraction of non-wild type residues at each position. For example, if the wild-type residue is A, then the library would contain mostly A with a mixture of C with T and G.
- Recombinant protein therapies
-
Therapies that are produced through recombinant DNA technology, which involves expressing and purifying a protein from bacterial or mammalian cells. Many biologics, such as monoclonal antibodies, are recombinant protein therapies.
- Phosphodiester linkage
-
The chemical linkage, connecting the monomeric units in a nucleic acid polymer. In general, the linkage takes the form RO(O)P(O−)OR′.
- Induced-fit
-
A biochemistry model in which the initial interactions of an enzyme–substrate complex (or antibody–target complex) are strengthened by conformational changes that increase the strength of the intermolecular interactions involved in substrate or target recognition.
- Fluorescent reporter
-
Molecules that elicit a fluorescent signal.
- Fluorescence resonance energy transfer
-
A photophysical effect in which energy is transferred between fluorescent or light-sensitive moieties.
- Mirror-image symmetry
-
Symmetry with respect to a reflection or plane of symmetry. Mirror-image molecules are non-superimposable. For example, your right hand cannot be superimposed on top of your left hand.
- von Willebrand factor
-
A glycoprotein found in the blood that is involved in haemostasis (blood clotting).
- Click chemistry
-
A general class of high-yielding cycloaddition reactions that occur between azide and alkyne functional groups. Click chemistry is often used to add new functional groups to biomolecules.
- Primer extension
-
A DNA (or xeno-nucleic acid (XNA)) replication step in which a nucleic acid primer is extended by annealing the primer to a template and copying the template with a polymerase.
- Fluorescence-activated cell sorting
-
A development of flow cytometry that enables sorting of a mixture of cells into two or more fractions based on the light scattering and fluorescence signal of each cell.
- Enzyme-linked immunosorbent assay
-
A diagnostic test that uses affinity reagents, typically antibodies, to detect a substance.
- Emulsion PCR
-
A variant of the polymerase chain reaction (PCR) that is performed in water-in-oil droplets.
Rights and permissions
About this article
Cite this article
Dunn, M., Jimenez, R. & Chaput, J. Analysis of aptamer discovery and technology. Nat Rev Chem 1, 0076 (2017). https://doi.org/10.1038/s41570-017-0076
Published:
DOI: https://doi.org/10.1038/s41570-017-0076