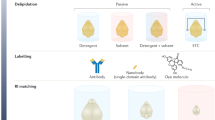
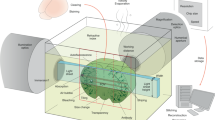
Abstract
The visualization of whole organs and organisms through tissue clearing and fluorescence volumetric imaging has revolutionized the way we look at biological samples. Its application to solid tumours is changing our perception of tumour architecture, revealing signalling networks and cell interactions critical in tumour progression, and provides a powerful new strategy for cancer diagnostics. This Review introduces the latest advances in tissue clearing and three-dimensional imaging, examines the challenges in clearing epithelia — the tissue of origin of most malignancies — and discusses the insights that tissue clearing has brought to cancer research, as well as the prospective applications to experimental and clinical oncology.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Spalteholz, W. Über das Durchsichtigmachen von menschlichen und tierischen Präparaten und seine theoretischen Bedingungen [German] (S. Hirzel, 1914).
Huisken, J., Swoger, J., Del Bene, F., Wittbrodt, J. & Stelzer, E. H. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 305, 1007–1009 (2004).
Voie, A. H., Burns, D. H. & Spelman, F. A. Orthogonal-plane fluorescence optical sectioning: three-dimensional imaging of macroscopic biological specimens. J. Microsc. 170, 229–236 (1993).
Ueda, H. R. et al. Tissue clearing and its applications in neuroscience. Nat. Rev. Neurosci. 21, 61–79 (2020).
Tedeschi, A. et al. Cep55 promotes cytokinesis of neural progenitors but is dispensable for most mammalian cell divisions. Nat. Commun. 11, 1746 (2020).
Dodt, H. U. et al. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat. Methods 4, 331–336 (2007).
Renier, N. et al. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 159, 896–910 (2014).
Cabeza-Cabrerizo, M. et al. Tissue clonality of dendritic cell subsets and emergency DCpoiesis revealed by multicolor fate mapping of DC progenitors. Sci. Immunol. 4, eaaw1941 (2019).
Tanaka, N. et al. Whole-tissue biopsy phenotyping of three-dimensional tumours reveals patterns of cancer heterogeneity. Nat. Biomed. Eng. 1, 796–806 (2017). This study shows clearing of FFPE samples.
Kubota, S. I. et al. Whole-body profiling of cancer metastasis with single-cell resolution. Cell Rep. 20, 236–250 (2017). This study quantifies body-wide metastasis by whole mouse clearing.
Garofalo, S. et al. Enriched environment reduces glioma growth through immune and non-immune mechanisms in mice. Nat. Commun. 6, 6623 (2015). This paper describes quantitative imaging of the lesion number and volume using tissue clearing.
Pan, C. et al. Deep learning reveals cancer metastasis and therapeutic antibody targeting in the entire body. Cell 179, 1661–1676.e19 (2019). This paper describes deep learning-based segmentation of metastasis in whole mice.
Oshimori, N., Oristian, D. & Fuchs, E. TGF-β promotes heterogeneity and drug resistance in squamous cell carcinoma. Cell 160, 963–976 (2015). This paper identifies the EMT-inducing niche by tissue clearing.
Messal, H. A. et al. Antigen retrieval and clearing for whole-organ immunofluorescence by FLASH. Nat. Protoc. 16, 239–262 (2021).
Lagerweij, T. et al. Optical clearing and fluorescence deep-tissue imaging for 3D quantitative analysis of the brain tumor microenvironment. Angiogenesis 20, 533–546 (2017).
Kingston, B. R., Syed, A. M., Ngai, J., Sindhwani, S. & Chan, W. C. W. Assessing micrometastases as a target for nanoparticles using 3D microscopy and machine learning. Proc. Natl Acad. Sci. USA 116, 14937–14946 (2019).
Susaki, E. A. & Ueda, H. R. Whole-body and whole-organ clearing and imaging techniques with single-cell resolution: toward organism-level systems biology in mammals. Cell Chem. Biol. 23, 137–157 (2016).
Richardson, D. S. & Lichtman, J. W. Clarifying tissue clearing. Cell 162, 246–257 (2015).
Tainaka, K., Kuno, A., Kubota, S. I., Murakami, T. & Ueda, H. R. Chemical principles in tissue clearing and staining protocols for whole-body cell profiling. Annu. Rev. Cell Dev. Biol. 32, 713–741 (2016).
Ueda, H. R. et al. Whole-brain profiling of cells and circuits in mammals by tissue clearing and light-sheet microscopy. Neuron 106, 369–387 (2020).
Azaripour, A. et al. A survey of clearing techniques for 3D imaging of tissues with special reference to connective tissue. Prog. Histochem. Cytochem. 51, 9–23 (2016).
Lloyd-Lewis, B. Multidimensional imaging of mammary gland development: a window into breast form and function. Front. Cell Dev. Biol. 8, 203 (2020).
Gomez-Gaviro, M. V., Sanderson, D., Ripoll, J. & Desco, M. Biomedical applications of tissue clearing and three-dimensional imaging in health and disease. iScience 23, 101432 (2020).
Susaki, E. A. et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell 157, 726–739 (2014).
Susaki, E. A. et al. Advanced CUBIC protocols for whole-brain and whole-body clearing and imaging. Nat. Protoc. 10, 1709–1727 (2015).
Tainaka, K. et al. Whole-body imaging with single-cell resolution by tissue decolorization. Cell 159, 911–924 (2014).
Sylwestrak, E. L., Rajasethupathy, P., Wright, M. A., Jaffe, A. & Deisseroth, K. Multiplexed intact-tissue transcriptional analysis at cellular resolution. Cell 164, 792–804 (2016).
Park, Y. G. et al. Protection of tissue physicochemical properties using polyfunctional crosslinkers. Nat. Biotechnol. 37, 73–83 (2018).
Zhao, S. et al. Cellular and molecular probing of intact human organs. Cell 180, 796–812.e19 (2020).
Ke, M. T., Fujimoto, S. & Imai, T. SeeDB: a simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat. Neurosci. 16, 1154–1161 (2013).
Ke, M. T. et al. Super-resolution mapping of neuronal circuitry with an index-optimized clearing agent. Cell Rep. 14, 2718–2732 (2016).
Kuwajima, T. et al. ClearT: a detergent- and solvent-free clearing method for neuronal and non-neuronal tissue. Development 140, 1364–1368 (2013).
Hama, H. et al. ScaleS: an optical clearing palette for biological imaging. Nat. Neurosci. 18, 1518–1529 (2015).
Pan, C. et al. Shrinkage-mediated imaging of entire organs and organisms using uDISCO. Nat. Methods 13, 859–867 (2016).
Jing, D. et al. Tissue clearing of both hard and soft tissue organs with the PEGASOS method. Cell Res. 28, 803–818 (2018).
Schwarz, M. K. et al. Fluorescent-protein stabilization and high-resolution imaging of cleared, intact mouse brains. PLoS ONE 10, e0124650 (2015).
Qi, Y. et al. FDISCO: advanced solvent-based clearing method for imaging whole organs. Sci. Adv. 5, eaau8355 (2019).
Hama, H. et al. Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat. Neurosci. 14, 1481–1488 (2011).
Hofmann, J., Gadjalova, I., Mishra, R., Ruland, J. & Keppler, S. J. Efficient tissue clearing and multi-organ volumetric imaging enable quantitative visualization of sparse immune cell populations during inflammation. Front. Immunol. 11, 599495 (2020).
Erturk, A. et al. Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nat. Protoc. 7, 1983–1995 (2012).
Erturk, A. et al. Three-dimensional imaging of the unsectioned adult spinal cord to assess axon regeneration and glial responses after injury. Nat. Med. 18, 166–171 (2012).
Tainaka, K. et al. Chemical landscape for tissue clearing based on hydrophilic reagents. Cell Rep. 24, 2196–2210.e9 (2018).
Messal, H. A. et al. Tissue curvature and apicobasal mechanical tension imbalance instruct cancer morphogenesis. Nature 566, 126–130 (2019). This paper identifies epithelial geometry-driven tumour initiation and progression by tissue clearing.
Chung, K. et al. Structural and molecular interrogation of intact biological systems. Nature 497, 332–337 (2013).
Chen, L. et al. UbasM: an effective balanced optical clearing method for intact biomedical imaging. Sci. Rep. 7, 12218 (2017).
Chi, J. et al. Three-dimensional adipose tissue imaging reveals regional variation in beige fat biogenesis and PRDM16-dependent sympathetic neurite density. Cell Metab. 27, 226–236.e3 (2018).
Li, W., Germain, R. N. & Gerner, M. Y. Multiplex, quantitative cellular analysis in large tissue volumes with clearing-enhanced 3D microscopy (Ce3D). Proc. Natl Acad. Sci. USA 114, E7321–E7330 (2017).
Treweek, J. B. et al. Whole-body tissue stabilization and selective extractions via tissue-hydrogel hybrids for high-resolution intact circuit mapping and phenotyping. Nat. Protoc. 10, 1860–1896 (2015).
Minsky, M. Microscopy apparatus. US Patent 3,013,467 (1961).
Sharpe, J. et al. Optical projection tomography as a tool for 3D microscopy and gene expression studies. Science 296, 541–545 (2002).
d’Esposito, A. et al. Computational fluid dynamics with imaging of cleared tissue and of in vivo perfusion predicts drug uptake and treatment responses in tumours. Nat. Biomed. Eng. 2, 773–787 (2018).
Lloyd-Lewis, B. et al. Imaging the mammary gland and mammary tumours in 3D: optical tissue clearing and immunofluorescence methods. Breast Cancer Res. 18, 127 (2016).
Li, S., Gestl, S. A. & Gunther, E. J. A multistage murine breast cancer model reveals long-lived premalignant clones refractory to parity-induced protection. Cancer Prev. Res. 13, 173–184 (2020).
Wei, M. et al. Volumetric chemical imaging by clearing-enhanced stimulated Raman scattering microscopy. Proc. Natl Acad. Sci. USA 116, 6608–6617 (2019).
Reynaud, E. G., Krzic, U., Greger, K. & Stelzer, E. H. Light sheet-based fluorescence microscopy: more dimensions, more photons, and less photodamage. HFSP J. 2, 266–275 (2008).
Herbert, S. P. et al. Arterial-venous segregation by selective cell sprouting: an alternative mode of blood vessel formation. Science 326, 294–298 (2009).
Power, R. M. & Huisken, J. A guide to light-sheet fluorescence microscopy for multiscale imaging. Nat. Methods 14, 360–373 (2017).
Sabdyusheva Litschauer, I. et al. 3D histopathology of human tumours by fast clearing and ultramicroscopy. Sci. Rep. 10, 17619 (2020).
Glaser, A. K. et al. Light-sheet microscopy for slide-free non-destructive pathology of large clinical specimens. Nat. Biomed. Eng. 1, 0084 (2017). This study develops 3D haematoxylin and eosin-like staining.
Tian, T., Yang, Z. & Li, X. Tissue clearing technique: recent progress and biomedical applications. J. Anat. 238, 489–507 (2021).
Tomer, R., Ye, L., Hsueh, B. & Deisseroth, K. Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nat. Protoc. 9, 1682–1697 (2014).
Belle, M. et al. Tridimensional visualization and analysis of early human development. Cell 169, 161–173.e12 (2017).
Matsumoto, K. et al. Advanced CUBIC tissue clearing for whole-organ cell profiling. Nat. Protoc. 14, 3506–3537 (2019).
Murakami, T. C. et al. A three-dimensional single-cell-resolution whole-brain atlas using CUBIC-X expansion microscopy and tissue clearing. Nat. Neurosci. 21, 625–637 (2018).
Chakraborty, T. et al. Light-sheet microscopy of cleared tissues with isotropic, subcellular resolution. Nat. Methods 16, 1109–1113 (2019).
Petran, M., Hadravský, M., Egger, M. D. & Galambos, R. Tandem-scanning reflected-light microscope. J. Optical Soc. Am. 58, 661–664 (1968).
Jonkman, J. & Brown, C. M. Any way you slice it — a comparison of confocal microscopy techniques. J. Biomol. Tech. 26, 54–65 (2015).
Wu, Y. et al. Resonant scanning with large field of view reduces photobleaching and enhances fluorescence yield in STED microscopy. Sci. Rep. 5, 14766 (2015).
Denk, W., Strickler, J. H. & Webb, W. W. Two-photon laser scanning fluorescence microscopy. Science 248, 73–76 (1990).
Lin, P. Y., Peng, S. J., Shen, C. N., Pasricha, P. J. & Tang, S. C. PanIN-associated pericyte, glial, and islet remodeling in mice revealed by 3D pancreatic duct lesion histology. Am. J. Physiol. Gastrointest. Liver Physiol. 311, G412–G422 (2016).
Lay, K. et al. Stem cells repurpose proliferation to contain a breach in their niche barrier. eLife 7, e41661 (2018).
Fiore, V. F. et al. Mechanics of a multilayer epithelium instruct tumour architecture and function. Nature 585, 433–439 (2020). This paper identifies early tumour architecture in complex epithelia by tissue clearing.
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Liebmann, T. et al. Three-dimensional study of Alzheimer’s disease hallmarks using the iDISCO clearing method. Cell Rep. 16, 1138–1152 (2016).
Casoni, F. et al. Development of the neurons controlling fertility in humans: new insights from 3D imaging and transparent fetal brains. Development 143, 3969–3981 (2016).
Belle, M. et al. A simple method for 3D analysis of immunolabeled axonal tracts in a transparent nervous system. Cell Rep. 9, 1191–1201 (2014).
Breckwoldt, M. O. et al. Correlated magnetic resonance imaging and ultramicroscopy (MR-UM) is a tool kit to assess the dynamics of glioma angiogenesis. eLife 5, e11712 (2016).
McIlwain, H. & Bachelard, H. S. Biochemistry and the Central Nervous System (Churchill Livingstone, 1985).
Oldham, M., Sakhalkar, H., Oliver, T., Allan Johnson, G. & Dewhirst, M. Optical clearing of unsectioned specimens for three-dimensional imaging via optical transmission and emission tomography. J. Biomed. Opt. 13, 021113 (2008).
Sung, K. et al. Simplified three-dimensional tissue clearing and incorporation of colorimetric phenotyping. Sci. Rep. 6, 30736 (2016).
Cancer Research UK. Types of cancer. Cancer Research UK https://www.cancerresearchuk.org/what-is-cancer/how-cancer-starts/types-of-cancer (2020).
Fu, Y. Y. et al. Microtome-free 3-dimensional confocal imaging method for visualization of mouse intestine with subcellular-level resolution. Gastroenterology 137, 453–465 (2009).
Liu, Y. A. et al. 3-D imaging, illustration, and quantitation of enteric glial network in transparent human colon mucosa. Neurogastroenterol. Motil. 25, e324–e338 (2013).
Bernier-Latmani, J. & Petrova, T. V. High-resolution 3D analysis of mouse small-intestinal stroma. Nat. Protoc. 11, 1617–1629 (2016).
Liu, Y. A. et al. Perivascular interstitial cells of cajal in human colon. Cell Mol. Gastroenterol. Hepatol. 1, 102–119 (2015).
Davis, F. M. et al. Single-cell lineage tracing in the mammary gland reveals stochastic clonal dispersion of stem/progenitor cell progeny. Nat. Commun. 7, 13053 (2016).
Rios, A. C. et al. Intraclonal plasticity in mammary tumors revealed through large-scale single-cell resolution 3D imaging. Cancer Cell 35, 618–632.e6 (2019). This paper identifies the EMT-inducing niche by tissue clearing.
Cai, R. et al. Panoptic imaging of transparent mice reveals whole-body neuronal projections and skull-meninges connections. Nat. Neurosci. 22, 317–327 (2019).
Gur-Cohen, S. et al. Stem cell-driven lymphatic remodeling coordinates tissue regeneration. Science 366, 1218–1225 (2019).
Tang, S. C. et al. Pancreatic neuro-insular network in young mice revealed by 3D panoramic histology. Diabetologia 61, 158–167 (2018).
Hong, S. M. et al. Three-dimensional visualization of cleared human pancreas cancer reveals that sustained epithelial-to-mesenchymal transition is not required for venous invasion. Mod. Pathol. 33, 639–647 (2020).
Gradauer, K. et al. Interaction with mixed micelles in the intestine attenuates the permeation enhancing potential of alkyl-maltosides. Mol. Pharm. 12, 2245–2253 (2015).
Hu, H. et al. Long-term expansion of functional mouse and human hepatocytes as 3D organoids. Cell 175, 1591–1606.e19 (2018).
Sachs, N. et al. Long-term expanding human airway organoids for disease modeling. EMBO J. 38, e100300 (2019).
Dekkers, J. F. et al. High-resolution 3D imaging of fixed and cleared organoids. Nat. Protoc. 14, 1756–1771 (2019).
Grist, S. M., Nasseri, S. S., Poon, T., Roskelley, C. & Cheung, K. C. On-chip clearing of arrays of 3-D cell cultures and micro-tissues. Biomicrofluidics 10, 044107 (2016).
van Royen, M. E. et al. Three-dimensional microscopic analysis of clinical prostate specimens. Histopathology 69, 985–992 (2016).
Noe, M. et al. Immunolabeling of cleared human pancreata provides insights into three-dimensional pancreatic anatomy and pathology. Am. J. Pathol. 188, 1530–1535 (2018).
Guldner, I. H. et al. An integrative platform for three-dimensional quantitative analysis of spatially heterogeneous metastasis landscapes. Sci. Rep. 6, 24201 (2016).
Brown, M. et al. Lymph node blood vessels provide exit routes for metastatic tumor cell dissemination in mice. Science 359, 1408–1411 (2018).
Pereira, E. R. et al. Lymph node metastases can invade local blood vessels, exit the node, and colonize distant organs in mice. Science 359, 1403–1407 (2018).
Song, E. et al. Optical clearing based cellular-level 3D visualization of intact lymph node cortex. Biomed. Opt. Express 6, 4154–4164 (2015).
Nojima, S. et al. CUBIC pathology: three-dimensional imaging for pathological diagnosis. Sci. Rep. 7, 9269 (2017).
von Neubeck, B. et al. An inhibitory antibody targeting carbonic anhydrase XII abrogates chemoresistance and significantly reduces lung metastases in an orthotopic breast cancer model in vivo. Int. J. Cancer 143, 2065–2075 (2018).
Yang, R. et al. The combination of two-dimensional and three-dimensional analysis methods contributes to the understanding of glioblastoma spatial heterogeneity. J. Biophotonics 13, e201900196 (2020).
Liu, Y. A. et al. 3-D visualization and quantitation of microvessels in transparent human colorectal carcinoma [corrected]. PLoS ONE 8, e81857 (2013).
Tanaka, N. et al. Mapping of the three-dimensional lymphatic microvasculature in bladder tumours using light-sheet microscopy. Br. J. Cancer 118, 995–999 (2018).
Mendler, C. T. et al. Tumor uptake of anti-CD20 Fabs depends on tumor perfusion. J. Nucl. Med. 57, 1971–1977 (2016).
Dobosz, M., Ntziachristos, V., Scheuer, W. & Strobel, S. Multispectral fluorescence ultramicroscopy: three-dimensional visualization and automatic quantification of tumor morphology, drug penetration, and antiangiogenic treatment response. Neoplasia 16, 1–13 (2014).
Poschinger, T. et al. Dynamic contrast-enhanced micro-computed tomography correlates with 3-dimensional fluorescence ultramicroscopy in antiangiogenic therapy of breast cancer xenografts. Invest. Radiol. 49, 445–456 (2014).
Si, Y. et al. Multidimensional imaging provides evidence for down-regulation of T cell effector function by MDSC in human cancer tissue. Sci. Immunol. 4, aaw9159 (2019). This paper identifies neutrophil hotspots that induce immune evasion by tissue clearing.
Chen, Y. et al. Three-dimensional imaging and quantitative analysis in CLARITY processed breast cancer tissues. Sci. Rep. 9, 5624 (2019).
Lee, S. S., Bindokas, V. P. & Kron, S. J. Multiplex three-dimensional optical mapping of tumor immune microenvironment. Sci. Rep. 7, 17031 (2017).
Brown, A. S. et al. Histologic changes associated with false-negative sentinel lymph nodes after preoperative chemotherapy in patients with confirmed lymph node-positive breast cancer before treatment. Cancer 116, 2878–2883 (2010).
Yokota, T. et al. Accuracy of preoperative diagnosis of lymph node metastasis for thoracic esophageal cancer patients from JCOG9907 trial. Int. J. Clin. Oncol. 21, 283–288 (2016).
Frechet, B., Kazakov, J., Thiffault, V., Ferraro, P. & Liberman, M. Diagnostic accuracy of mediastinal lymph node staging techniques in the preoperative assessment of nonsmall cell lung cancer patients. J. Bronchol. Interv. Pulmonol. 25, 17–24 (2018).
King, C. R. & Long, J. P. Prostate biopsy grading errors: a sampling problem? Int. J. Cancer 90, 326–330 (2000).
Catalona, W. J., Stein, A. J. & Fair, W. R. Grading errors in prostatic needle biopsies: relation to the accuracy of tumor grade in predicting pelvic lymph node metastases. J. Urol. 127, 919–922 (1982).
Epstein, J. I. Prostate cancer grading: a decade after the 2005 modified system. Mod. Pathol. 31, S47–S63 (2018).
Ahdoot, M. et al. MRI-targeted, systematic, and combined biopsy for prostate cancer diagnosis. N. Engl. J. Med. 382, 917–928 (2020).
Bostrom, P. J. et al. Staging and staging errors in bladder cancer. Eur. Urol. Suppl. 9, 2–9 (2010).
Kruskal, J. B., Kane, R. A., Sentovich, S. M. & Longmaid, H. E. Pitfalls and sources of error in staging rectal cancer with endorectal us. Radiographics 17, 609–626 (1997).
Torres, R., Vesuna, S. & Levene, M. J. High-resolution, 2- and 3-dimensional imaging of uncut, unembedded tissue biopsy samples. Arch. Pathol. Lab. Med. 138, 395–402 (2014).
Yang, B. et al. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell 158, 945–958 (2014).
Hsueh, B. et al. Pathways to clinical CLARITY: volumetric analysis of irregular, soft, and heterogeneous tissues in development and disease. Sci. Rep. 7, 5899 (2017).
Tanaka, N. et al. Three-dimensional single-cell imaging for the analysis of RNA and protein expression in intact tumour biopsies. Nat. Biomed. Eng. 4, 875–888 (2020).
Sun, D. E. et al. Click-ExM enables expansion microscopy for all biomolecules. Nat. Methods 18, 107–113 (2021).
Liu, J. T. C. et al. Harnessing non-destructive 3D pathology. Nat. Biomed. Eng. 5, 203–218 (2021).
Eisenstein, M. Transparent tissues bring cells into focus for microscopy. Nature 564, 147–149 (2018).
Franca, C. M. et al. 3D-imaging of whole neuronal and vascular networks of the human dental pulp via CLARITY and light sheet microscopy. Sci. Rep. 9, 10860 (2019).
Hook, P. et al. Whole blood clot optical clearing for nondestructive 3D imaging and quantitative analysis. Biomed. Opt. Express 8, 3671–3686 (2017).
Bulantova, J. et al. Trichobilharzia regenti (Schistosomatidae): 3D imaging techniques in characterization of larval migration through the CNS of vertebrates. Micron 83, 62–71 (2016).
Kang, G. Y., Rhyu, H. J., Choi, H. H., Shin, S. J. & Hyun, Y. M. 3D Imaging of the transparent Mycobacterium tuberculosis-infected lung verifies the localization of innate immune cells with granuloma. Front. Cell Infect. Microbiol. 10, 226 (2020).
Zaeck, L. M. et al. 3D reconstruction of SARS-CoV-2 infection in ferrets emphasizes focal infection pattern in the upper respiratory tract. Preprint at bioRxiv https://doi.org/10.1101/2020.10.17.339051 (2020).
Wang, H. et al. Deep learning enables cross-modality super-resolution in fluorescence microscopy. Nat. Methods 16, 103–110 (2019).
Murray, E. et al. Simple, scalable proteomic imaging for high-dimensional profiling of intact systems. Cell 163, 1500–1514 (2015).
Hou, B. et al. Scalable and DiI-compatible optical clearance of the mammalian brain. Front. Neuroanat. 9, 19 (2015).
Cai, R. et al. Panoptic vDISCO imaging reveals neuronal connectivity, remote trauma effects and meningeal vessels in intact transparent mice. Preprint at bioRxiv https://doi.org/10.1101/374785 (2018).
Masselink, W. et al. Broad applicability of a streamlined ethyl cinnamate-based clearing procedure. Development 146, dev166884 (2019).
Hahn, C. et al. High-resolution imaging of fluorescent whole mouse brains using stabilised organic media (sDISCO). J. Biophotonics 12, e201800368 (2019).
Tseng, S. J. et al. Integration of optical clearing and optical sectioning microscopy for three-dimensional imaging of natural biomaterial scaffolds in thin sections. J. Biomed. Opt. 14, 044004 (2009).
Liu, K. et al. Metabolic stress drives sympathetic neuropathy within the liver. Cell Metab. 33, 666–675.e4 (2021).
Chen, F., Tillberg, P. & Boyden, E. Expansion microscopy. Science 347, 543–548 (2015).
Tillberg, P. W. et al. Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nat. Biotechnol. 34, 987–992 (2016).
Miyabayashi, K. et al. Intraductal transplantation models of human pancreatic ductal adenocarcinoma reveal progressive transition of molecular subtypes. Cancer Discov. 10, 1566–1589 (2020).
Li, W., Germain, R. N. & Gerner, M. Y. High-dimensional cell-level analysis of tissues with Ce3D multiplex volume imaging. Nat. Protoc. 14, 1708–1733 (2019).
Greenbaum, A. et al. Bone CLARITY: clearing, imaging, and computational analysis of osteoprogenitors within intact bone marrow. Sci. Transl. Med. 9, eaah6518 (2017).
van Ineveld, R. et al. Revealing the spatio-phenotypic patterning of cells in healthy and tumor tissues with mLSR-3D and STAPL-3D. Nat. Biotechnol. https://doi.org/10.1038/s41587-021-00926-3 (2021).
Kugler, R. et al. Novel imaging of the prostate reveals spontaneous gland contraction and excretory duct quiescence together with different drug effects. FASEB J. 32, 1130–1138 (2018).
LeCun, Y., Bengio, Y. & Hinton, G. Deep learning. Nature 521, 436–444 (2015).
Todorov, M. I. et al. Machine learning analysis of whole mouse brain vasculature. Nat. Methods 17, 442–449 (2020).
Berg, S. et al. ilastik: interactive machine learning for (bio)image analysis. Nat. Methods 16, 1226–1232 (2019).
Schoppe, O. et al. Deep learning-enabled multi-organ segmentation in whole-body mouse scans. Nat. Commun. 11, 5626 (2020).
Isensee, F., Jaeger, P. F., Kohl, S. A. A., Petersen, J. & Maier-Hein, K. H. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 18, 203–211 (2021).
Sullivan, D. P. et al. Deep learning is combined with massive-scale citizen science to improve large-scale image classification. Nat. Biotechnol. 36, 820–828 (2018).
McKinney, S. M. et al. International evaluation of an AI system for breast cancer screening. Nature 577, 89–94 (2020).
Savage, N. How AI is improving cancer diagnostics. Nature 579, S14–S16 (2020).
Paluch, E. K. After the greeting: realizing the potential of physical models in cell biology. Trends Cell Biol. 25, 711–713 (2015).
Acknowledgements
The authors are grateful to K. Ng (The Francis Crick Institute), R. Ng (University of British Columbia), A. Neumann (The Francis Crick Institute and Imperial College London) and C. Basier (The Francis Crick Institute) for critical reading of the manuscript. This work was supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001039, FC001317), the UK Medical Research Council (FC001039, FC001317), the Wellcome Trust (FC001039, FC001317), the European Molecular Biology Organization (EMBO long-term fellowship ALTF 452-2019 to H.A.M.) and the Doctor Josef Steiner Foundation (to J.v.R.).
Author information
Authors and Affiliations
Contributions
J.A., H.A.M. and M.Z.T. researched data for the article, made substantial contribution to discussion of content and wrote, reviewed and edited the manuscript before submission. A.B. and J.v.R. made substantial contribution to discussion of content and wrote, reviewed and edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
H.A.M. and A.B. are inventors on a UK patent application (1818567.8) on clearing solutions for tissue clearing and three-dimensional (3D) imaging. The other authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Cancer thanks Ali Erturk, who co-reviewed with Chenchen Pan, Hiroki Ueda and Per Uhlén for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Aivia 3D: https://www.aivia-software.com/aivia-3d
Imaris: https://imaris.oxinst.com
Vision4D: https://imaging.arivis.com/en/imaging-science/arivis-vision4d
Supplementary information
Glossary
- Light-sheet fluorescence microscopy
-
(LSFM). A fluorescence microscopy technique in which only a plane of the sample is illuminated at one time.
- Numerical aperture
-
A dimensionless number proportional to the refractive index of the medium and the sine of the maximum half-angle of light passing through a lens.
- Raman scattering microscopy
-
A technique to image chemical bonds in biological samples by quantum amplification via stimulated emission.
- Point scanning confocal microscopes
-
Fluorescence microscopes that illuminate a single point of the sample and use a pinhole to eliminate the out of focus signal.
- Axially swept light-sheet microscopy
-
A light-sheet fluorescence microscopy technique in which illumination is scanned in its direction of propagation.
- Free working distance
-
The distance from the front of the objective to the closest surface of the sample in focus.
- Spinning disk confocal microscopy
-
A confocal microscopy technique that uses multiple pinholes on a spinning disk to direct the excitation and emission light beams and increase the imaging speed.
- Resonant scanning confocal microscopy
-
A confocal microscopy technique with galvanometric mirrors for fast image acquisition.
- Vibratome
-
An instrument that slices micrometre-scale sample sections with a vibrating blade.
- Expansion microscopy
-
(ExM). A sample preparation protocol in which specimens are isometrically swollen in a polymer gel to image small structures.
- Click chemistry
-
A biocompatible reaction allowing covalent bonding of a biomolecule to a substrate of choice.
Rights and permissions
About this article
Cite this article
Almagro, J., Messal, H.A., Zaw Thin, M. et al. Tissue clearing to examine tumour complexity in three dimensions. Nat Rev Cancer 21, 718–730 (2021). https://doi.org/10.1038/s41568-021-00382-w
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41568-021-00382-w
This article is cited by
-
Beyond genetics: driving cancer with the tumour microenvironment behind the wheel
Nature Reviews Cancer (2024)
-
Emerging strategies to investigate the biology of early cancer
Nature Reviews Cancer (2024)
-
Matrigel-based organoid culture of malignant mesothelioma reproduces cisplatin sensitivity through CTR1
BMC Cancer (2023)
-
In focus in HCB
Histochemistry and Cell Biology (2023)
-
Three-dimensional visualization of human brain tumors using the CUBIC technique
Brain Tumor Pathology (2023)