Abstract
During clathrin-mediated endocytosis, dozens of proteins assemble into an interconnected network at the plasma membrane. As initiators of endocytosis, Eps15 and Fcho1/2 concentrate downstream components, while permitting dynamic rearrangement within the budding vesicle. How do initiator proteins meet these competing demands? Here we show that Eps15 and Fcho1/2 rely on weak, liquid-like interactions to catalyse endocytosis. In vitro, these weak interactions promote the assembly of protein droplets with liquid-like properties. To probe the physiological role of these liquid-like networks, we tuned the strength of initiator protein assembly in real time using light-inducible oligomerization of Eps15. Low light levels drove liquid-like assemblies, restoring normal rates of endocytosis in mammalian Eps15-knockout cells. By contrast, initiator proteins formed solid-like assemblies upon exposure to higher light levels, which stalled vesicle budding, probably owing to insufficient molecular rearrangement. These findings suggest that liquid-like assembly of initiator proteins provides an optimal catalytic platform for endocytosis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 /Â 30Â days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data supporting this work are available on request from the corresponding author. Source data are provided with this paper.
Code availability
Data analysis was conducted using an ImageJ FRAP profiler plugin (http://worms.zoology.wisc.edu/ImageJ/FRAP_Profiler_v2.java) and CMEanalysis software (https://www.utsouthwestern.edu/labs/danuser/software/). No custom code was generated for this study.
References
Henne, W. M. et al. FCHo proteins are nucleators of clathrin-mediated endocytosis. Science 328, 1281–1284 (2010).
Taylor, M. J., Perrais, D. & Merrifield, C. J. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 9, e1000604 (2011).
Wang, L., Johnson, A., Hanna, M. & Audhya, A. Eps15 membrane-binding and-bending activity acts redundantly with Fcho1 during clathrin-mediated endocytosis. Mol. Biol. Cell 27, 2675–2687 (2016).
Sengar, A. S., Wang, W., Bishay, J., Cohen, S. & Egan, S. E. The EH and SH3 domain Ese proteins regulate endocytosis by linking to dynamin and Eps15. EMBO J. 18, 1159–1171 (1999).
Mayers, J. R. et al. Regulation of ubiquitin-dependent cargo sorting by multiple endocytic adaptors at the plasma membrane. Proc. Natl Acad. Sci. USA 110, 11857–11862 (2013).
Ma, L. et al. Transient Fcho1/2⋅Eps15/R⋅AP-2 nanoclusters prime the AP-2 clathrin adaptor for cargo binding. Dev. Cell 37, 428–443 (2016).
Morgan, J. R., Prasad, K., Jin, S., Augustine, G. J. & Lafer, E. M. Eps15 homology Domain–NPF motif interactions regulate clathrin coat assembly during synaptic vesicle recycling. J. Biol. Chem. 278, 33583–33592 (2003).
Chen, H. et al. Epsin is an EH-domain-binding protein implicated in clathrin-mediated endocytosis. Nature 394, 793–797 (1998).
Cupers, P., Haar, Eter, Boll, W. & Kirchhausen, T. Parallel dimers and anti-parallel tetramers formed by epidermal growth factor receptor pathway substrate clone 15 (EPS15). J. Biol. Chem. 272, 33430–33434 (1997).
Henne, W. M. et al. Structure and analysis of FCHo2 F-BAR domain: a dimerizing and membrane recruitment module that effects membrane curvature. Structure 15, 839–852 (2007).
Lu, R. & Drubin, D. G. Selection and stabilization of endocytic sites by Ede1, a yeast functional homologue of human Eps15. Mol. Biol. Cell 28, 567–575 (2017).
Tebar, F., Sorkina, T., Sorkin, A., Ericsson, M. & Kirchhausen, T. Eps15 is a component of clathrin-coated pits and vesicles and is located at the rim of coated pits. J. Biol. Chem. 271, 28727–28730 (1996).
Sochacki, K. A., Dickey, A. M., Strub, M.-P. & Taraska, J. W. Endocytic proteins are partitioned at the edge of the clathrin lattice in mammalian cells. Nat. Cell Biol. 19, 352–361 (2017).
Carbone, R. et al. eps15 and eps15R are essential components of the endocytic pathway. Cancer Res. 57, 5498–5504 (1997).
Benmerah, A., Bayrou, M., Cerf-Bensussan, N. & Dautry-Varsat, A. Inhibition of clathrin-coated pit assembly by an Eps15 mutant. J. Cell Sci. 112, 1303–1311 (1999).
Benmerah, A., Poupon, V., Cerf-Bensussan, N. & Dautry-Varsat, A. Mapping of Eps15 domains involved in its targeting to clathrin-coated pits. J. Biol. Chem. 275, 3288–3295 (2000).
Santonico, E., Panni, S., Falconi, M., Castagnoli, L. & Cesareni, G. Binding to DPF-motif by the POB1 EH domain is responsible for POB1–Eps15 interaction. BMC Biochem. 8, 29 (2007).
Simonsen, A. C. & Bagatolli, L. A. Structure of spin-coated lipid films and domain formation in supported membranes formed by hydration. Langmuir 20, 9720–9728 (2004).
Wei, M.-T. et al. Phase behaviour of disordered proteins underlying low density and high permeability of liquid organelles. Nat. Chem. 9, 1118–1125 (2017).
Elbaum-Garfinkle, S. et al. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl Acad. Sci. USA 112, 7189–7194 (2015).
Banjade, S. & Rosen, M. K. Phase transitions of multivalent proteins can promote clustering of membrane receptors. eLife Sci. 3, e04123 (2014).
Li, P. et al. Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340 (2012).
Shin, Y. & Brangwynne, C. P. Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 (2017).
Milovanovic, D., Wu, Y., Bian, X. & Camilli, P. D. A liquid phase of synapsin and lipid vesicles. Science 361, 604–607 (2018).
Alberti, S., Gladfelter, A. & Mittag, T. Considerations and challenges in studying liquid–liquid phase separation and biomolecular condensates. Cell 176, 419–434 (2019).
Hein, M. Y. et al. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell 163, 712–723 (2015).
Bugaj, L. J., Choksi, A. T., Mesuda, C. K., Kane, R. S. & Schaffer, D. V. Optogenetic protein clustering and signaling activation in mammalian cells. Nat. Methods 10, 249–252 (2013).
Shin, Y. et al. Spatiotemporal control of intracellular phase transitions using light-activated optoDroplets. Cell 168, 159–171 (2017).
Aguet, F. et al. Membrane dynamics of dividing cells imaged by lattice light-sheet microscopy. Mol. Biol. Cell 27, 3418–3435 (2016).
Grimm, J. B. et al. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat. Methods 12, 244–250 (2015).
Loerke, D. et al. Cargo and dynamin regulate clathrin-coated pit maturation. PLoS Biol. 7, e1000057 (2009).
Ehrlich, M. et al. Endocytosis by random initiation and stabilization of clathrin-coated pits. Cell 118, 591–605 (2004).
Boulant, S., Kural, C., Zeeh, J.-C., Ubelmann, F. & Kirchhausen, T. Actin dynamics counteract membrane tension during clathrin-mediated endocytosis. Nat. Cell Biol. 13, 1124–1131 (2011).
Dambournet, D. et al. Genome-edited human stem cells expressing fluorescently labeled endocytic markers allow quantitative analysis of clathrin-mediated endocytosis during differentiation. J. Cell Biol. 217, 3301–3311 (2018).
Kaksonen, M. & Roux, A. Mechanisms of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 19, 313–326 (2018).
Mettlen, M., Chen, P.-H., Srinivasan, S., Danuser, G. & Schmid, S. L. Regulation of clathrin-mediated endocytosis. Annu. Rev. Biochem. 87, 871–896 (2018).
Avinoam, O., Schorb, M., Beese, C. J., Briggs, J. A. G. & Kaksonen, M. Endocytic sites mature by continuous bending and remodeling of the clathrin coat. Science 348, 1369–1372 (2015).
Zhuo, Y. et al. Dynamic interactions between clathrin and locally structured elements in a disordered protein mediate clathrin lattice assembly. J. Mol. Biol. 404, 274–290 (2010).
Kozak, M. & Kaksonen, M. Phase separation of Ede1 promotes the initiation of endocytic events. Preprint at bioRxiv https://doi.org/10.1101/861203 (2019).
Smith, S. M., Baker, M., Halebian, M. & Smith, C. J. Weak molecular interactions in clathrin-mediated endocytosis. Front. Mol. Biosci. 4, 72 (2017).
Huang, W. Y. C. et al. Phosphotyrosine-mediated LAT assembly on membranes drives kinetic bifurcation in recruitment dynamics of the Ras activator SOS. Proc. Natl Acad. Sci. USA 113, 8218–8223 (2016).
Chen, Y. et al. Dynamic instability of clathrin assembly provides proofreading control for endocytosis. J. Cell Biol. 218, 3200–3211 (2019).
Gellman, A. J. Oligomer desorption during heterogeneous catalytic synthesis of polymers. Catal. Today 105, 144–151 (2005).
Bickford, L. C., Mossessova, E. & Goldberg, J. A structural view of the COPII vesicle coat. Curr. Opin. Struct. Biol. 14, 147–153 (2004).
Sato, K. & Nakano, A. Mechanisms of COPII vesicle formation and protein sorting. FEBS Lett. 581, 2076–2082 (2007).
Sandvig, K., Kavaliauskiene, S. & Skotland, T. Clathrin-independent endocytosis: an increasing degree of complexity. Histochem. Cell Biol. 150, 107–118 (2018).
Kennedy, M. J. et al. Rapid blue-light-mediated induction of protein interactions in living cells. Nat. Methods 7, 973–975 (2010).
Zeno, W. F., Rystov, A., Sasaki, D. Y., Risbud, S. H. & Longo, M. L. Crowding-Induced mixing behavior of lipid bilayers: Examination of mixing energy, phase, packing geometry, and reversibility. Langmuir 32, 4688–4697 (2016).
Ran, F. A. et al. Genome engineering using the CRISPR–Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Aguet, F., Antonescu, C. N., Mettlen, M., Schmid, S. L. & Danuser, G. Advances in analysis of low signal-to-noise images link dynamin and AP2 to the functions of an endocytic checkpoint. Dev. Cell 26, 279–291 (2013).
Acknowledgements
We thank T. Kirchhausen for the gift of SUM159/AP-2σ2-HaloTag cells and L. Lavis for the gift of JF646 HaloTag ligand. This research was supported by the National Institutes of Health through grants R35GM139531 and R01GM112065 to J.C.S. and E.M.L., grant R01GM118933 to E.M.L., and F32GM133138 to K.J.D., by a National Science Foundation Graduate Research Fellowship (DGE-1610403) to G.K, and by a grant from the Welch Foundation (F-2047) to J.C.S. The University of Texas Health Science Center at San Antonio (UTHSCSA) Center for Macromolecular Interactions is supported by the Cancer Therapy and Research Center through the National Cancer Institute P30 Grant CA054174, and Texas State funds provided through the UTHSCSA Office of the Vice President for Research.
Author information
Authors and Affiliations
Contributions
K.J.D., E.M.L., G.K., C.C.H., and J.C.S. designed experiments. K.J.D. and J.C.S. wrote and edited the manuscript. K.J.D., G.K., J.B.R., L.W., C.C.H., E.M.L., and J.C.S. performed experiments and analysed data. All authors consulted on manuscript preparation and editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Cell Biology thanks Volker Haucke and the other, anonymous, reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Fcho2 assembles into protein-rich domains with Eps15 on membranes and in solution.
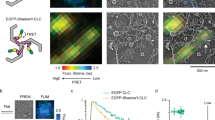
a-e, Fcho2 is labeled with Atto-594. Eps15 contains an N-terminal 6xHis tag and is labeled with CF488a. a-b, Center slices (upper panels) and corresponding z-projections (lower panels) of representative GUVs incubated with 500 nM of the indicated protein(s). GUVs contain 79% DOPC, 15% DOPS, 5% PtdIns(4,5)P2, and 1% DPEG10-biotin. a, Full-length Fcho2 alone decorates GUVs homogeneously. b, GUVs incubated with both Fcho2 and Eps15 display protein-rich domains. c, 7 μM Fcho2 clusters into small, irregular aggregates. d, When combined at a 1:34 ratio, Fcho2 and Eps15 colocalize in protein droplets. e, Time course of a fusion event between droplets containing Fcho2 (magenta) and Eps15 (green). Scale bars are 5 μm in a-c, e and 10 µm in d.
Extended Data Fig. 2 Eps15 and Fcho1 co-localize in protein-rich domains on membrane surfaces.
Extended Data Fig. 3 Eps15/Fcho1 droplet:solution partitioning decreases at increasing temperature.
a-c, Representative images of protein droplets at increasing temperatures, followed by the return to room temperature after heat removal. Plots show fluorescence intensity of Eps15-CF488a measured along dotted lines in each image. Intensity is normalized to the maximum value in the first 25 °C panel for each set of images. Fcho1 is unlabeled. Total protein concentration is held at 7 μM while Fcho1:Eps15 ratio is varied. Droplets are formed from (a) 7 μM Eps15, (b) 0.1 μM Fcho1, 6.9 μM Eps15, and (c) 0.2 μM Fcho1, 6.8 μM Eps15. See Source Data Extended Data Fig. 3.
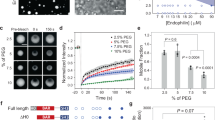
Extended Data Fig. 4 Eps15 mutants and Fcho1 assemble to varying degrees in solution.
a-c, Fcho1 is labeled with Atto-594, Eps15 mutants are labeled with CF488a. Panels on the left show 7 μM Eps15 mutant alone, set of panels on the right show 6.8 μM Eps15 mutant combined with 0.2 μM Fcho1 (34:1). Cartoons depict binding interaction between Fcho1 and Eps15 mutants. a, Eps15 lacking the EH domains (Eps15-∆3xEH) does not form droplets on its own, but when combined with Fcho1 forms small droplets. b, Eps15 lacking the C-terminal disordered domain (Eps15-∆CTD) does not form droplets on its own and addition of Fcho1 does not induce droplet formation, reinforcing that the CTD of Eps15 mediates its interaction with Fcho1. c, Eps15 containing mutated Fcho1-binding DPF motifs (amino acids 623-636; Eps15-DPF>APA) robustly assembles into droplets on its own and co-assembles into droplets with Fcho1, presumably because the disordered domains of Eps15 and Fcho1 interact even in the absence of 3 key DPF motifs. Scale bar is 10 µm.
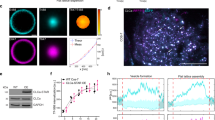
Extended Data Fig. 5 Controls for Eps15-Cry2 cell experiments.
a, Left: whole cell lysates from WT SUM159/AP-2-HaloTag cells and WT SUM159/AP-2-HaloTag cells gene edited by CRISPR to disrupt Eps15 were separated by SDS-PAGE and immunoblotted for Eps15 and GAPDH. Right: whole cell lysates from WT SUM159/AP-2-HaloTag cells and WT SUM159/AP-2-HaloTag cells transfected with siRNA against Eps15R were collected 24 hours post-transfection. Proteins were separated by SDS-PAGE and immunoblotted for Eps15R and GAPDH. b, Itsn1-mCherry (upper panels), Fcho1-mCherry (lower panels), Eps15-Cry2-GFP, and AP2-HaloTag conjugated to JF646 colocalize in Eps15∆ cells expressing Eps15-Cry2 and exposed to low blue light levels. White arrowheads indicate examples of colocalization in endocytic structures. Notably, Fcho1-mCherry/Eps15-Cry2-GFP co-expression often resulted in the formation of large, persistent aggregates on the plasma membrane, denoted by yellow arrowheads. Scale bar is 5 µm. c, The lifetime distributions of AP-2 σ2-HaloTag-labeled endocytic structures in Eps15 knockout cells expressing Eps15-mCherry and in wild-type Eps15 cells are nearly identical. d, The average plasma membrane fluorescence intensity of AP-2 σ2-HaloTag::JF646 and Eps15-mCherry in the first frame of each movie analysed in a and Fig. 4. Eps15∆ n=10 biologically independent cell samples, 25,269 pits. WT Eps15 n=10 biologically independent cell samples, 8,969 pits. No light n=11 biologically independent cell samples, 21,996 pits. Low light n=17 biologically independent cell samples, 14,222 pits. Strong light n=12 biologically independent cell samples, 13,978 pits. e, Lifetime distributions of clathrin-coated structures in Eps15∆/Eps15R knockdown cells expressing Eps15-Cry2 at no, low, or strong blue light exposure. Plots show frequency of short-lived (<20 s, magenta), productive (20-180 s, gray), and long-lived (>180 s, yellow) structures for each condition. No blue light exposure resulted in 42 ± 3% (SEM) of CCPs being short-lived (<20 s). Low blue light exposure significantly reduced the frequency of short-lived CCPs from 42 ± 3% to 36 ± 4% (t-test, p=0.042, n=5, 5). While 2-3% of pits were long-lived (>180 s) in cells exposed to no or low blue light, the frequency of long-lived pits increased significantly to 6 ± 1% (t-test, p=0.009, n=5, 5) in cells exposed to strong blue light. No light n=5 biologically independent cell samples, 31,427 pits. Low light n=5 biologically independent cell samples, 27,026 pits. Strong light n=5 biologically independent cell samples, 17,623 pits. f, The lifetime distributions of AP-2 σ2-HaloTag-labeled endocytic structures in Eps15 knockout cells expressing Eps15-mCherry exposed to either no blue light or 50 µW “strong†blue light are nearly identical. g Plot from Fig. 6f and (h) plot from Fig. 6h displaying the individual data points that were averaged together for each FRAP curve. n=5-6 biologically independent samples. Data are presented as mean ± SEM. See Source Data Extended Data Fig. 5.
Extended Data Fig. 6 The assembly state of initiator proteins impacts CME dynamics.
When Eps15 (green) and Fcho1 (magenta) exist in an unassembled, or dilute phase on the membrane surface, abortive structures are favored. In productive structures, Eps15 and Fcho1 assemble into a liquid protein phase capable of exchange with molecules in solution. Further assembly of Eps15 and Fcho1 into a gel or solid phase limits molecular exchange and promotes stalled endocytic structures.
Supplementary information
Supplementary Video 1
Eps15–Fcho1-rich protein domains merge together on multibilayers. Video of frames shown in Fig. 1h. 100 nM Atto594-labelled Fcho1 (magenta) and 100 nM CF488a-labelled Eps15 (green) were applied to a multibilayer consisting of 73% DOPC, 25% DOPS, and 2% DOGS–NTA-Ni and imaged immediately. Arrowheads indicate merging protein-rich domains. Interval between frames is 3 s, scale bar is 2 μm.
Supplementary Video 2
Droplets consisting of Eps15 only or Eps15 and Fcho1 merge together, whereas Fcho1 droplets do not merge. Video of frames shown in Fig. 2b, f. Solutions of 7 μM CF488a-labelled Eps15 (upper left), 7 μM Atto594-labelled Fcho1 (lower left), or 6.8 μM CF488a-labelled Eps15 and 0.2 μM dark Fcho1 (right) were prepared. Interval between frames is 0.5 s for Eps15 only and 1 s for Fcho1 only and Eps15–Fcho1, scale bar is 2 μm.
Supplementary Video 3
Eps15 droplets dissolve uniformly when heated and reform when cooled. Video on the left shows 7 μM CF488a-labelled Eps15 as it was heated from 30 °C to 33 °C at a rate of approximately 1 °C min−1. Video on the right shows 7 μM CF488a-labelled Eps15 as it was allowed to cool from 33 °C to 30 °C at a rate of approximately 2 °C min−1. Interval between frames is 1 s, scale bar is 10 μm.
Supplementary Video 4
Addition of Fcho1 to a single-phase solution of Eps15 induces droplet formation. Video corresponding to Fig. 2h. 3 μM CF488a-labelled Eps15 (green) does not form phase-separated droplets. 0.12 μM Atto594-labelled Fcho1 (magenta) was added at time 0 s and resulted in the formation of droplets that incorporated Eps15 within seconds. Focal plane is near the coverslip, where droplets gradually settled after forming in solution. Video begins 30 s after Fcho1 addition, interval between frames is 1 s, scale bar is 5 μm.
Supplementary Video 5
Droplets consisting of Eps15–Cry2 and Eps15 show light-dependent fusion behaviour. Video of frames shown in Figs. 3c, 5a. A solution of 3 μM CF488a-labelled Eps15 (green) and 1 μM Atto594-labelled Eps15–Cry2 (magenta) was exposed to either low blue light (left) or strong blue light (right). Droplets displayed liquid-like merging behaviour under low blue light, but failed to merge under strong blue light, indicating solid-like assembly. Interval between frames is 2 s, scale bar is 5 μm.
Supplementary Video 6
Blue-light exposure impacts the dynamics of clathrin-coated structures in cells expressing Eps15–Cry2. Representative movies of cells used for analysis in Figs. 4 and 5. Cells were gene-edited to delete endogenous Eps15 and to express AP-2 σ2-HaloTag. AP-2 is labelled with JF646 dye and displayed in cyan. Cells express either no Eps15 (Eps15Δ), Eps15–mCherry (WT Eps15) or Eps15–Cry2–mCherry (Light off, Low light and Strong light), displayed in red. Movies begin after 1 min of imaging with or without blue light. Interval between frames is 3 s, scale bar is 10 μm.
Source data
Source Data Fig. 1
Statistical source data for Fig. 1
Source Data Fig. 2
Statistical source data for Fig. 2
Source Data Fig. 3
Statistical source data for Fig. 3
Source Data Fig. 4
Statistical source data for Fig. 4
Source Data Fig. 5
Statistical source data for Fig. 5
Source Data Fig. 6
Statistical source data for Fig. 6
Source Data Extended Data Fig. 2
Statistical source data for Extended Data Fig. 2
Source Data Extended Data Fig. 3
Statistical source data for Extended Data Fig. 3
Source Data Extended Data Fig. 5
Statistical source data for Extended Data Fig. 5
Source Data Extended Data Fig. 5
Unprocessed western blots for Extended Data Fig. 5
Rights and permissions
About this article
Cite this article
Day, K.J., Kago, G., Wang, L. et al. Liquid-like protein interactions catalyse assembly of endocytic vesicles. Nat Cell Biol 23, 366–376 (2021). https://doi.org/10.1038/s41556-021-00646-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-021-00646-5