Abstract
Nuclear factor erythroid 2-related factor 2 (NRF2) hyperactivation has been established as an oncogenic driver in a variety of human cancers, including non-small cell lung cancer (NSCLC). However, despite massive efforts, no specific therapy is currently available to target NRF2 hyperactivation. Here, we identify peptidylprolyl isomerase A (PPIA) is required for NRF2 protein stability. Ablation of PPIA promotes NRF2 protein degradation and blocks NRF2-driven growth in NSCLC cells. Mechanistically, PPIA physically binds to NRF2 and blocks the access of ubiquitin/Kelch Like ECH Associated Protein 1 (KEAP1) to NRF2, thus preventing ubiquitin-mediated degradation. Our X-ray co-crystal structure reveals that PPIA directly interacts with a NRF2 interdomain linker via a trans-proline 174-harboring hydrophobic sequence. We further demonstrate that an FDA-approved drug, cyclosporin A (CsA), impairs the interaction of NRF2 with PPIA, inducing NRF2 ubiquitination and degradation. Interestingly, CsA interrupts glutamine metabolism mediated by the NRF2/KLF5/SLC1A5 pathway, consequently suppressing the growth of NRF2-hyperactivated NSCLC cells. CsA and a glutaminase inhibitor combination therapy significantly retard tumor progression in NSCLC patient-derived xenograft (PDX) models with NRF2 hyperactivation. Our study demonstrates that targeting NRF2 protein stability is an actionable therapeutic approach to treat NRF2-hyperactivated NSCLC.
Similar content being viewed by others
Introduction
The transcription factor NRF2 (nuclear factor erythroid 2-related factor 2) and its negative regulator KEAP1 (Kelch Like ECH Associated Protein 1) contribute profoundly to the maintenance of the cellular redox homeostasis1. Under oxidative and electrophilic stress, NRF2 is released from KEAP1 and translocated to the nucleus to initiate the transcription of the antioxidant program2. This transient and inducible response was recognized originally as a protective mechanism against tumorigenesis3. However, many studies have subsequently indicated that constitutive NRF2 activation exhibits potent pro-tumorigenic properties in many types of cancer4, including lung cancer.
Pan-cancer genome analysis of The Cancer Genome Atlas (TCGA) revealed that NRF2 activation was particularly enhanced in non-small cell lung cancer (NSCLC)5. Genomic alterations that activated the NRF2 pathway ranked as the third most common in lung adenocarcinoma (LUAD, 25%), largely due to loss-of-function mutations in KEAP16. Also, KRAS oncogenic mutations, the most prevalent oncogenic drivers in LUAD, potentiate transcriptional activation of NRF27. KEAP1 mutations co-exist in approximately 20% of KRAS-mutant LUAD8. Activated NRF2 not only promotes lung tumorigenesis through maintaining redox homeostasis and detoxification of reactive oxygen species (ROS)7, but also facilitates aggressive lung cancer via multiple pro-metastasis factors6,9. Collectively, these comprehensive studies have established abnormal NRF2 activation as a pivotal oncogenic driver in lung cancer, implying a potential benefit of targeting NRF2 in cancer therapy. However, targeted therapies that directly inhibit NRF2 remain inaccessible, emphasizing the necessity of exploring an alternative strategy.
In this study, we explore the innovative idea that NRF2 protein stability per se may (finally) enable a therapeutic approach for NRF2-hyperactivated NSCLC. Our biochemical, structural, and therapeutic model findings collectively reveal a proline-dependent regulatory mechanism for NRF2 stability, and we demonstrate that this mechanism can be pharmacologically targeted in NSCLC with NRF2 hyperactivation.
Results
Identification and validation of Cyclosporin A (CsA) as a chemical inducer of targeted NRF2 protein degradation
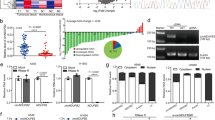
Given the known recalcitrance of directly inhibiting transcription factor NRF210, we adopted an alternative targeting strategy ‒ exploiting the protein degradation machineries of NRF2. We first investigated NRF2 protein expression in 17 NSCLC cell lines. We detected a significant increase of NRF2 protein levels in nuclear extracts of KEAP1 and/or KRAS mutant cell lines compared to corresponding wildtype (WT) cell lines (Table 1, Fig. 1A, B), which was concordant with previously studies11,12. To explore potential chemical inducer of NRF2 protein degradation, we generated a fluorescence-based sensor DsRed-IRES-EGFP-NRF2 (RIG-NRF2) for monitoring NRF2 stability in NSCLC cells (Fig. 1C). Briefly, we fused NRF2 to the fluorescent indicator protein EGFP and used DsRed as an internal control in a bicistronic lentiviral vector pLenti-DsRed-IRES-EGFP13,14. So, the EGFP/DsRed ratio can reflect the relative stability of NRF2. The resultant RIG-NRF2 was then stably transfected into NSCLC cell line A549 (Fig. 1C), and a monoclonal cell population was generated by limiting dilution.
A Representative immunoblot analysis of NRF2 protein levels in nuclear extracts of 17 NSCLC cell lines. B Dot plot showing the relative nuclear NRF2 protein levels in cell lines with KEAP1 and/or KRAS co-mutations (n = 9 cell lines) and in cell lines with KEAP1 and KRAS WT (n = 8 cell lines) related to Fig. 1A. C Overview of the DsRed-IRES-EGFP-NRF2 (RIG-NRF2) screen. CMV, CMV promoter; IRES, Internal ribosome entry site. D Flow cytometry analysis of DsRed and EGFP levels of A549-RIG-NRF2 cells treated with CHX (10 μM), MG132 (10 μM), Ki696 (1 μM), ML334 (50 μM) and CsA (10 μM). The gating strategy are provided in Supplementary Fig. 13. E IC50 values of CsA against a panel of NSCLC cell lines with distinct genetic profile. F Representative immunoblot analysis of NRF2 protein levels in a panel of NSCLC cells upon CsA treatment (10 μM). The results of panels (A, D and F) are representative of three independent experiments. E represents mean ± SD of three independent experiments. P value was analyzed using Two-tailed unpaired Student’s t-test, P < 0.05 was considered statistically significant. Source data are provided as a Source Data file.
An in-house library of 805 compounds (including clinical or approved drugs) was screened to identify potential compounds that can regulate NRF2 protein stability using the RIG-NRF2-expressing cell line above. The percentage of the decreased level of NRF2 was calculated by comparing that of the vehicle control (0%) with that of the cycloheximide (CHX)-treated group (100%) (Fig. 1D). Two known NRF2 activators Ki696 and ML334 were used as controls15,16. This screen revealed that the macrocyclic peptide drug cyclosporin A (CsA) exerted the strongest NRF2 degradation promoting effect among the library drugs (Fig. 1D and Supplementary Fig. 1A). We next examined CsA’s effects on endogenous NRF2 in two representative NRF2-hyperactivated NSCLC cell lines (A549 and H2030). Immunoblotting analysis showed that CsA treatment reduced the NRF2 protein level in both cell lines (Supplementary Fig. 1B, C). In contrast, Q-PCR analysis revealed that mRNA level of NRF2 was not affected by CsA treatment (Supplementary Fig. 1D). The reduction of NRF2 protein level in CsA-treated cells was reversed by proteasome inhibition with MG132 (Supplementary Fig. 1E). CsA-treated cells exhibited a profound higher level of ubiquitinated NRF2 upon MG132 treatment, as compared with vehicle-treated cells (Supplementary Fig. 1F). These findings indicate that the ubiquitin-proteasome pathway is required for the degradation of NRF2 induced by CsA treatment. Intriguingly, we also observed a decrease of NRF2T80K, a gain-of-function mutant of NRF2, upon CsA treatment (Supplementary Fig. 1G).
We further found that CsA treatment significantly repressed NRF2-driven luciferase activity and inhibited the expression of two classical NRF2 target genes, heme oxygenase 1 (HMOX1) and NAD(P)H: quinone oxidoreductase 1 (NQO1) in NRF2-hyperactivated A549 and H2030 cells, but not in H1650 and H1975 cells with low basal NRF2 level (Supplementary Fig. 1H-1J). Together, these results demonstrate that the FDA-approved drug CsA can induce the decrease of NRF2 protein levels in NRF2-hyperactivated NSCLC cells.
CsA impedes NRF2-dependent growth of NSCLC cells
We next used CsA as a probe to delineate the functional contribution of NRF2 degradation in NSCLC. To this end, we monitored the potential anti-proliferative activities of CsA in NSCLC cell lines. Intriguingly, KEAP1/KRAS co-mutant cell lines, excepting for H1944, with augmented NRF2 protein levels were highly sensitive to CsA; KEAP1 or KRAS single mutant cell lines partially responded to CsA; however, the corresponding WT cell lines and EBC-1 cells harboring heterogeneous D77V mutation with a low basal NRF2 level were largely resistant to CsA treatment (Fig. 1E, Supplementary Fig. 2A, B, Supplementary Table 1). Correlated with these results, CsA treatment profoundly reduced NRF2 protein levels in NRF2-hyperactivated NSCLC cells (Fig. 1F). We further found that siRNA-mediated knockdown of NRF2 impaired the anti-proliferative capacity of CsA in NRF2-hyperactivated A549 and H2030 cells (Supplementary Fig. 2C). Overexpression of NRF2 or its gain-of function mutant (NRF2T80K) sensitized H1650 and H1975 cells, which have low basal NRF2 levels, to CsA treatment (Supplementary Fig. 2C). In line with these results, pharmacological activation of NRF2 by Ki696 and ML334 rendered sensitivity to CsA treatment in these two cell lines (Supplementary Fig. 2D). Together, these results demonstrate that CsA can suppress the NRF2-dependent growth and proliferation of NSCLC cells.
Peptidylprolyl isomerase A (PPIA) is required for CsA-induced reduction of NRF2 levels and NSCLC cell proliferation
Peptidylprolyl isomerase A (PPIA, or CypA) is a specific cytosolic binding protein for CsA and is responsible for CsA’s immunomodulating effects17. It has been shown to catalyze the cis-trans isomerization of proline residues and to regulate protein folding and stability18. These ideas led us to speculate that PPIA may be involved in the CsA-mediated promotion of NRF2 degradation. Pursuing this hypothesis, we knocked out (KO) the PPIA gene from A549 and H2030 using CRISPR/Cas9 genome editing. PPIA-knockout (PPIA-KO) resulted in profound reduction in the NRF2 protein levels, whereas showed no significant changes in mRNA levels of NRF2, in both KO cell lines (Fig. 2A and Supplementary Fig. 3A, B). However, CsA treatment caused no reduction in the NRF2 protein levels in the PPIA-KO cells (Fig. 2B and Supplementary Fig. 3C). We have demonstrated that CsA can induce NRF2 degradation via ubiquitin-proteasome system (Supplementary Fig. 1E, F). In line with this observation, PPIA-KO cells have a profound increased level of ubiquitinated NRF2 upon MG132 treatment compared with the unedited control cells (Fig. 2C and Supplementary Fig. 3D). These results demonstrate that PPIA is required for CsA-triggered degradation of the NRF2 protein.
A Representative immunoblot analysis of NRF2 protein levels in PPIA-WT or PPIA-KO A549 cells. Quantitative data was provided in the right panel. B Representative immunoblot analysis of NRF2 protein levels in PPIA-WT or PPIA KO A549 cells treated with CsA (0, 3, 10 μM) for 48 h. Quantitative data was provided in the right panel. C Immunoprecipitation of NRF2 followed by immunoblot analysis with anti-ubiquitin antibody detected the NRF2 ubiquitination in PPIA-WT and PPIA-KO A549 cells. D CHX chase assay of NRF2 protein stability in PPIA-WT or PPIA-KO A549 cells. Cells were treated with CHX (100 μg/mL) at the indicated time points and then subjected to immunoblot. Quantitative data was provided in the right panel. E Representative immunoblot results demonstrate that PPIA WT, but not PPIA catalytically dead PPIA variant (PPIAR55A&F60A), can restore the NRF2 level in PPIA-KO A549 cells. Quantitative data was provided in the right panel. F Schematic diagram of NanoBRET™ Ubiquitination Assay using the HaloTag®-Ubiquitin and NRF2-NanoLuc® fusion construct. G BRET signal of HEK293T cells transfected with HaloTag®-Ubiquitin and NRF2-Nanoluc® following siPPIA treatment or not. H Immunoprecipitation of NRF2 followed by immunoblot analysis with anti-ubiquitin antibody detected the NRF2 ubiquitination in HEK293T following siPPIA treatment or not. I Relative cell growth of A549 upon PPIA KO or CsA (10 μM) treatment in the presence or absence of NRF2 overexpression. Quantitative results were shown in upper panel and representative colony image was presented in lower panel. C and H are representative of three independent experiments. (A–E), G and (I) represent mean ± SD of three independent experiments. P values were analyzed using Two-tailed unpaired Student’s t-test, P < 0.05 was considered statistically significant. Source data are provided as a Source Data file.
We next monitored NRF2 stability in WT and PPIA-KO A549 and H2030 cells by pre-treatment with cycloheximide (CHX) to block de novo protein synthesis. As a result, NRF2 protein half-life was much shorter in the PPIA-KO cells (A549, t1/2 = 38.3 ± 0.7 min; H2030, t1/2 = 30.1 ± 1.3 min) compared with the unedited control cells (A549, t1/2 = 78.6 ± 1.7 min; H2030, t1/2 = 61.2 ± 2.8 min) (Fig. 2D and Supplementary Fig. 3E). Moreover, we have additionally examined the NRF2 stability in two NRF2-hyperactivated NSCLC cell lines (H460 and H2122). To do this, we knocked out (KO) the PPIA gene from H460 and H2122 using CRISPR/Cas9 technology. Consistently, NRF2 protein half-life was much shorter in the PPIA-KO cells (H460, t1/2 = 44.5 ± 1.4 min; H2122, t1/2 = 36.9 ± 0.5 min) compared with the unedited control cells (H460, t1/2 > 80 min; H2122, t1/2 > 80 min) (Supplementary Fig. 3F, G). Implicating PPIA’s cis-trans isomerization activity on proline, we also overexpressed WT PPIA or a catalytically dead PPIA in the PPIA-KO background cells (R55A&F60A, retaining less than 1% enzymatic catalytic activity)19. Cells expressing WT PPIA, but not catalytically dead PPIA, displayed significantly elevated NRF2 levels (Fig. 2E and Supplementary Fig. 3H), indicating that catalytically competent PPIA is required for protecting NRF2 from degradation.
To further delineate the PPIA’s protection effect on NRF2 from degradation by ubiquitin-proteasome system, we thus established a live-cell ubiquitin assay, NanoBRET™ Ubiquitination Assay, which can sensitively detect ubiquitination on a target protein (Promega). NanoBRET™ Ubiquitination Assay was performed using the HaloTag®-Ubiquitin and NRF2-NanoLuc® fusion proteins (Fig. 2F). As shown in Fig. 2G, a significantly increase in the NanoBRET™ signal was observed for NRF2/Ubiquitin in HEK293T upon siPPIA treatment, compared with the HEK293T cells upon siControl treatment. In line with these results, siPPIA treatment profoundly elevated the NRF2 ubiquitination as detected by immunoblotting (Fig. 2H). In contrast, siPPIA treatment failed to elevate the ubiquitination of NRF2 K7/R7 mutant (seven lysines in Neh2 domain of NRF2 required for NRF2 ubiquitination were all mutant into arginine)20,21 (Supplementary Fig. 4A, B). Moreover, overexpression of WT PPIA, but not catalytically dead PPIA (R55A&F60A), displayed significantly reduced NanoBRET™ signal of NRF2/Ubiquitin and NRF2 ubiquitination by immunoblotting (Supplementary Fig. 4C, D). NRF2 ubiquitination is mediated by its negative regulator KEAP1, a component of an E3 ubiquitin ligase complex, upon binding22. We then asked whether PPIA affected the interaction of KEAP1 and NRF2. To this end, we performed co-immunoprecipitation analysis and we observed that overexpression of PPIA profoundly impaired the binding of KEAP1 to NRF2 (Supplementary Fig. 4E). These results suggest that PPIA can protect NRF2 from ubiquitination.
Next, we sought to establish a functional link between PPIA and NRF2. We found that firstly, NRF2-driven luciferase expression was markedly decreased in PPIA-KO A549 and H2030 cells and the inhibitory activities of CsA on ARE-reporter assay was abolished by PPIA-KO (Supplementary Fig. 4F). Secondly, CsA inhibits the mRNA expression of NRF2 target genes HMOX1 and NQO1 in a PPIA-dependent manner (Supplementary Fig. 4G). Thirdly, PPIA-KO or CsA treatment can significantly impair the colony formations in A549 and H2030 cells, but not in H1650 and H1975 cells with low basal NRF2 levels; overexpression of NRF2 in A549 and H2030 cells confers resistance to PPIA-KO and CsA treatment (Fig. 2I, Supplementary Fig. 4H,I). Finally, we employed A549 cells with inducible shCtrl or shPPIA to generate tumors in mice. Doxycycline (20 mg/kg) was orally administrated to induce shRNA expression. Of note, PPIA knockdown strongly reduced growth of A549 xenograft tumors. In contrast, H1650 cells with inducible shCtrl or shPPIA showed comparable tumor growth capacity (Supplementary Fig. 4J). Taken together, these results demonstrate that PPIA plays a vital role in protecting NRF2 protein from CsA-mediated degradation and consequently enhancing NRF2’s transcriptional and proliferative functions in NSCLC cells.
PPIA interacts with NRF2
Next, we investigated whether PPIA interacts with NRF2. Co-immunoprecipitation results revealed an interaction between PPIA and NRF2 using A549 cell lysates (Fig. 3A). Moreover, we found that PPIA’s enzymatic activity is required for the PPIA-NRF2 interaction, since no interaction was observed for a catalytically dead PPIA variant (PPIAR55A&F60A) (Fig. 3B). In addition, pull-down analysis using recombinant PPIA-conjugated beads showed that CsA profoundly blocked the interaction of PPIA and NRF2 (Fig. 3C). Collectively, these results indicate that PPIA can bind to NRF2.
A Co-immunoprecipitation analysis of endogenous proteins of PPIA and NRF2 using A549 cell lysates. B Pull-down assay of PPIA WT/NRF2 and PPIAR55A&F60A/NRF2. A549 cell lysate was incubated with PPIA WT or PPIAR55A&F60A pre-loaded beads, and NRF2 protein retained on the beads were detected by immunoblot. C The influence of CsA on the interaction between PPIA and NRF2. A549 cell lysate were incubated with PPIA pre-loaded beads in the presence of CsA (0, 3, 10 μM) and then subjected to pull-down assay. D Schematic diagram of various NRF2 truncation. E The binding of full-length and truncated NRF2 to PPIA as determined by pull-down assay. Various NRF2 truncations were overexpressed in HEK293T cells (as described in D). F CHX chase assay of NRF2 (WT or mutant P174A) protein stability. Cells were treated with CHX (100 μg/mL) at the indicated time points and then subjected to immunoblot (upper panel). Quantitative data was provided in the lower panel. G Crystal structure of PPIA in complex with NRF2 fragment PPIA-Binding Motif (PBM) (169VAQVAPVD176). PBM peptide is deeply embedded into catalytic pocket of PPIA presented as gray surface. 172VAPV175 residues of NRF2 peptide are displayed as slate stick. H Expansion of the catalytic pocket of PPIA in complex with NRF2 fragment PBM. PPIA is shown in gray cartoon and the interacted residues are displayed as salmon sticks. NRF2 PBM fragment is presented as slate sticks. The black dashed line denotes the hydrogen contact. The results of panels (A–C, E, F) are representative of three independent experiments. F represents mean ± SD of three independent experiments. P values were analyzed using Two-tailed unpaired Student’s t-test, P < 0.05 was considered statistically significant. Source data are provided as a Source Data file.
The complex crystal structure reveals that proline 174 (P174) of NRF2 is essential for PPIA binding
We then generated truncation variants to map the interaction of NRF2 and PPIA according to NRF2’s functional NRF2-ECH homology (Neh) domains (Fig. 3D). Pull-down using recombinant PPIA-conjugated beads showed that the intrinsically disordered linker between the Neh4 and Neh5 domains (135-182aa), but not the Neh domains, is required for PPIA binding (Fig. 3E). Given the proline cis−trans isomerization activity of PPIA, we suspected possible involvement of proline residues in the PPIA/NRF2 interaction. Pursuing this, we mutated each of the five prolines in the flexible linker region (P145, P148, P154, P165, and P174) of NRF2 individually. Among these five prolines, P174A abolished the binding of NRF2 to PPIA in pull-down assay (Supplementary Fig. 5A). Correlated with this result, this NRF2 P174A mutant was less stable with a shorten half-life of 35.3 ± 1.4 min than NRF2 WT (t1/2 = 52.3 ± 3.7 min) (Fig. 3F). The NRF2 P174A mutant also elevated NanoBRET™ signal of NRF2/Ubiquitin and NRF2 ubiquitination by immunoblotting, suggesting a crucial role of PPIA in regulating NRF2 stability (Supplementary Fig. 5B, C).
We next sought to characterize the structural features of P174-dependent NRF2 and PPIA interaction. We synthesized a Proline-174-harboring hydrophobic peptide sequence (169VAQVAPVD176) located in the linker between the Neh4 and Neh5 domains and co-crystalized it with full-length PPIA using a hanging-drop apparatus. The structure of the complex was resolved with a resolution of 1.81 Å, and a final Rfree of 0.2478 (Table 2, Supplementary Table 2). Overall, the structural data demonstrated that the VAPV fragment deeply embedded in the catalytic pocket of PPIA formed by R55, F60, M61, D66, F67, N101, and F113 (Fig. 3G, H). The guanidine of PPIA’s R55 side chain directly interacts with the main chain of P174 of the NRF2 peptide. The amidogen and carboxyl of the PPIA’s N102 main chain respectively form hydrogen bonds with A173 and V175 of the NRF2 peptide (Fig. 3H). Of note, P174 is buried in the catalytic cleft of PPIA in a trans conformation, which may be the result of the peptide bonds preceding and following P174, tightly anchoring P174 in the active pocket of PPIA. Intriguingly, the hydrophobic sequence motif 169VAQVAPVD176 is highly conserved among primate species (Supplementary Fig. 5D), representing a unique PPIA-Binding-Motif (PBM).
Next, we compared the structural differences between PPIA-PBM and PPIA-CsA (PDB ID: 1CWA)23. The two structures present some differences around the catalytic pocket, including the β3 strand formed by residues H54 to I56, the β4 strand formed by residues F60 to G64, and the α2 helix formed by residues E120 to D123 (Supplementary Fig. 5E). The most obvious variation is in the right-angle direction of guanidine in the R55 side chain (shifted by about 4.3 Å), leading to shifting of the β3 strand’s tail. A possible reason for this conformational change is strong interaction with VAPV that moves the R55 residue towards P174 in PBM fragment. The VAPV peptide displayed a similar conformation at this location as residues 3 to 6 of CsA (Supplementary Fig. 5E), which can potentially explain CsA’s disruption of the PPIA and NRF2 interaction.
We also employed a classic Nuclear Magnetic Resonance (NMR)-based approach to monitor the protein-peptide interaction. The 15N-edited heteronuclear single quantum coherence (15N-HSQC) spectrum revealed that the addition of the PBM peptide solution resulted in pronounced chemical shift perturbations. In line with the complex structure of PPIA and PBM fragment, a residue-specific NMR spectroscopy showed that C52, G59, D66, F67, and F113—all located in the catalytic pocket of PPIA—as the most affected residues (Supplementary Fig. 5F, G). Together, these results demonstrate that NRF2 via its unique PBM motif directly binds to PPIA and P174 is vital for this binding and maintaining NRF2 stability by PPIA.
PPIA inhibition impairs glutamine metabolism in NSCLC cells with NRF2 hyperactivation
Previous studies established that abnormal NRF2 activation promotes glutamine addiction in NSCLC24,25. We therefore evaluated the effect of PPIA inhibition on glutamine metabolism in A549 and H2030 cells. We found that glutamine supplementation significantly enhanced cell growth, whereas the growth-promoting effects of this supplementation were impeded upon PPIA KO and upon CsA treatment of both cell lines (Fig. 4A, Supplementary Fig. 7A). Glutamine is converted to glutamate by glutaminase and further oxidized by tricarboxylic acid (TCA) cycle in mitochondrial26. A549 and H2030 cells were loaded with minimal medium in which glutamate was used as sole carbon source. These two cell lines were subjected to oxygen consumption rate (OCR) assay via a Seahorse XFe96 analyzer. We found that PPIA KO and CsA treatment results in reduced basal and maximal OCR, suggesting impaired glutamine metabolism in mitochondria of both cell lines (Fig. 4B, Supplementary Fig. 7B). We further performed the glutamine metabolism tracing using 13C5-glutamine and LC-MS/MS (Supplementary Fig. 6A). As shown in Supplementary Fig. 6B, PPIA KO can significantly decrease the flux of glutamine-derived TCA metabolites, including glutamate, α-ketoglutarate, succinate, fumarate, and malate. Consistent with these observations, the supplementation of glutamate, α-ketoglutarate, succinate, and fumarate can significantly rescue the cell viability upon PPIA KO (Supplementary Fig. 6C).
A The cell viability of A549 cells upon CsA (10 μM) treatment or upon PPIA KO in the presence of glutamine (2, 1, 0.5 mM) was measured by MTT assay. Gln, glutamine. B Oxygen consumption rate (OCR) plotted over time in A549 cells following CsA (10 μM) treatment or PPIA KO. C Heatmap showing the mRNA levels of different glutamine membrane transporters in PPIA-WT or PPIA-KO A549 cells. D-E Q-PCR D and immunoblot E analysis of SLC1A5 in PPIA-WT or PPIA-KO A549 cells following NRF2 overexpression. F-G Q-PCR F and immunoblot G results of SLC1A5 in A549 cells treated with CsA (10 μM for 48 h) with or without NRF2 overexpression. H Relative glutamine levels in A549 cells upon CsA (10 μM) treatment or upon PPIA KO, following SLC1A5 or NRF2 overexpression. I A549 cells upon CsA (10 μM) treatment or upon PPIA KO following SLC1A5 overexpression were subjected to colony formation. Quantitative results were shown in upper panel and representative colony image was presented in lower panel. The results of panels (C, E, G) are representative of three independent experiments. A, B, D, F, H and I represent mean ± SD of three independent experiments. P values were analyzed using Two-tailed unpaired Student’s t-test, P < 0.05 was considered statistically significant. Source data are provided as a Source Data file.
We also examined whether PPIA affects glutamine uptake/transport by using Q-PCR to assess the transcription levels of human glutamine transporters (Fig. 4C). The largest decrease in transcription in PPIA-KO cells was the transcripts of the SLC1A5, a high-affinity glutamine transporter for maintaining influx of glutamine. We also observed a significant decrease in both the mRNA and protein levels of SLC1A5 upon PPIA KO and upon CsA treatment in both A549 and H2030 cells. Of note, overexpression of NRF2 reversed SLC1A5 expression loss (Fig. 4D–G, Supplementary Fig. 7C–F).
We next investigated the effects of PPIA inhibition on cellular glutamine content. As anticipated, PPIA KO and CsA treatment resulted in 68.42 ± 1.25% and 67.09 ± 4.10% reductions of intracellular glutamine content in A549 cells, respectively. Whereas, NRF2 overexpression effectively restored glutamine level (Fig. 4H). The similar results were also observed in H2030 cells (Supplementary Fig. 7G). To assess the functional role of SLC1A5 in glutamine transport, we overexpressed SLC1A5 in A549 and H2030 cells. As expected, we found that SLC1A5 overexpression rescued the reduction in glutamine content upon PPIA KO and upon CsA treatment in these two cell lines (Fig. 4H and Supplementary Fig. 7G). We also observed that SLC1A5 overexpression effectively rescued the cell growth inhibition in both A549 and H2030 cells upon PPIA KO and upon CsA treatment (Fig. 4I, Supplementary Fig. 7H). Taken together, these results suggest that PPIA inhibition may impede glutamine metabolism in a SLC1A5-dependent manner in NRF2-hyperactivated NSCLC cells.
NRF2 activates SLC1A5 transcription through its downstream gene KLF5
Immunoblotting against NRF2 and SLC1A5 in a panel of 17 NSCLC cell lines showed that these two proteins were commonly co-expressed (Supplementary Fig. 8A; Pearson R = 0.7271, P = 0.0009). RNA-sequencing data of NSCLC tissues derived from GEO database (GSE8894) also indicated a positive correlation between NRF2 and SLC1A5 (Supplementary Fig. 8B; Pearson R = 0.5197, P < 0.0001)27. We then examined the possibility that SLC1A5 may be a direct target gene of the NRF2 transcription factor by generating a luciferase reporter construct containing the SLC1A5 promoter (ranging from -2000 bp to +100 bp). Overexpression of NRF2 increased the promoter activity of SLC1A5, doing so in a dose-dependent manner (Supplementary Fig. 8C). To map the transcription activation region, we assessed a series of truncated SLC1A5 promoters and ultimately identified the binding region from -500 bp to -10 bp (Supplementary Fig. 8D). However, analysis using the JASPAR database28 did not detect any NRF2 binding sequence (ARE element) within this region, strongly suggesting that NRF2 per se is not responsible for the observed transcriptional activation of SLC1A5.
We therefore suspected an indirect regulation mechanism, so we looked for transcription factors common to i) the NRF2-regulated genes identified based on chromatin immunoprecipitation (ChIP)-Seq data29 and ii) predicted transcription factor binding to the promoter of SLC1A5 in the GeneCards database30 (Supplementary Fig. 8E). Among the 14 such genes, Kruppel Like Factor 5 (KLF5) exhibited the highest predicted JASPAR score (Supplementary Fig. 8F). Firstly, we examine whether KLF5 is a target gene of the NRF2. We found that NRF2 enhanced KLF5 transcription using a luciferase reporter vector containing the KLF5 promoter (ranging from -4000 bp to 0 bp) (Fig. 5A). Two potential NRF2 binding sequences (ARE element) within the KLF5 promoter (GGGACACAGCC, -3621 to -3611 bp and GTGTCACTGCA, -2068 to -2058 bp) (Fig. 5B) were predicted by JASPAR and confirmed by ChIP-PCR (Fig. 5C). Subsequently, we asked whether KLF5 induce SLC1A5 transcription. Luciferase reporter (the -500 bp to -10 bp region) analysis demonstrated that KLF5 was capable of inducing SLC1A5 transcription (Fig. 5D). ChIP-PCR results demonstrated that KLF5 binds to a specific motif (GCCCCGCCCC, -47 to -38 bp) within the SLC1A5 promoter (Fig. 5E, F).
A pGL3-Luc vector containing human KLF5 promoter (-4,000 to 0 bp) and pCDNA3.1-Flag-NRF2 were co-transfected into HEK293T cells with a ratio of 1:0.5 to 1:8 and luciferase activity was determined. B Analysis of NRF2 consensus motif enrichment in the KLF5 promoter (-4,000 to 0 bp) predicted by JASPAR database. Matched consensus motifs are shown in schematic. C ChIP-PCR analysis of the enrichment of NRF2 at the promoter region of KLF5 in A549 cells. D pGL3-Luc vector containing human SLC1A5 promoter (-500 to +10 bp) and pCDNA3.1-Flag-KLF5 were co-transfected into HEK293T cells with a ratio of 1:0.5 to 1:8 and luciferase activity was determined. E Analysis of KLF5 consensus motif enrichment in the SLC1A5 promoter predicted by JASPAR database. Matched consensus sequences are in bold. F ChIP-PCR analysis of the enrichment of KLF5 at the promoter region of SLC1A5 in A549 cells. G Q-PCR and immunoblot results of SLC1A5 in A549 cells treated with siNRF2 in the presence or absence of KLF5 overexpression. H Relative glutamine level in A549 cells following siNRF2 treatment in the presence or absence of KLF5 overexpression. I Colony formation of A549 cells treated with CsA or siNRF2 in the presence or absence of KLF5 overexpression. Quantitative results were shown in upper panel and representative colony image was presented in lower panel. J Correlation analysis of KLF5/NRF2 or KLF5/SLC1A5 gene expression in clinical NSCLC tumor samples (n = 138 samples). The data are derived from public dataset (GSE8894) and analyzed in PrognoScan. The results of panels (C, F, G) are representative of three independent experiments. A, C, D, F, G–I represent mean ± SD of three independent experiments. P values were analyzed using Two-tailed unpaired Student’s t-test, P < 0.05 was considered statistically significant. Source data are provided as a Source Data file.
Next, we observed that NRF2 overexpression increased the mRNA levels of KLF5, while siNRF2-mediated knockdown significantly reduced KLF5 accumulation (Fig. 5G and Supplementary Fig. 9A, B). Additionally, we found that the effects of NRF2 knockdown in A549 and H2030 cells—including decreased mRNA level and protein accumulation of SLC1A5 and decreased intracellular glutamine content—were restored upon KLF5 overexpression (Fig. 5G, H, Supplementary Fig. 9B, C). Moreover, KLF5 knockdown reduced the intracellular glutamine level and the colony formation capacity of A549 and H2030 cells, which were restored following SLC1A5 overexpression (Supplementary Fig. 9D,E). By contrast, KLF5 overexpression rescued the growth capacity of A549 and H2030 cells upon CsA treatment and NRF2 knockdown (Fig. 5I, Supplementary Fig. 9F). We further examined the co-expression relationship of NRF2, KLF5 and SLC1A5 in NSCLC. Quantitation of the immunoblotting showed protein expression of KLF5 is positively correlated with that of NRF2 or SLC1A5 in NSCLC cell lines (Supplementary Fig. 8A). Additionally, analysis of the human NSCLC dataset in the GEO database (GSE8894)27 suggested that the mRNA expression of KLF5 is positively associated with that of NRF2 or SLC1A5 (Fig. 5J). These results collectively demonstrate that NRF2-KLF5 signaling promotes the transcription activation of glutamine transporter SLC1A5 in NSCLC cells.
Co-targeting of PPIA and glutaminase potently inhibits tumor growth of NRF2-hyperactivated NSCLC
Glutamine metabolism is explored therapeutically as a new approach for the treatment of many types of cancers, including NSCLC31. Glutamine is transported into cells through plasma membrane transporters (e.g., SLC1A5) and is subsequently converted to glutamate by glutaminase to fuel cancer cell growth32. We next evaluated the anti-proliferation effects of a combination therapy comprising CsA and glutaminase inhibitor CB-839 against A549 and H2030. Co-treatment with CsA and CB-839 caused a pronounced reduction in the viability of both A549 and H2030 cells, with the co-treatment outperforming either of the single agents (Supplementary Fig. 10A). Combining CsA with CB-839 demonstrated a synergistic anti-proliferation effect with combination index (CI) values less than 1 at varying concentrations (Supplementary Fig. 10A). Notably, overexpression of KEAP1 profoundly reduced NRF2 protein levels and conferred resistance to CsA and CB839 combination treatment in both A549 and H2030 cells (Supplementary Fig. 10B, C). H1650 and H1975 cells with low NRF2 expression are insensitive to CsA/CB-839 co-treatment (Supplementary Fig. 10D). Note that a glutamine deprivation assay indicated that the anti-proliferation effect from the CsA and CB-839 drug combination was dependent on the presence of glutamine (Fig. 6A and Supplementary Fig. 10E). We also conducted 3D colony formation analysis with A549 and H2030 cells and observed a significant loss of colony formation capacity upon the CsA and CB-839 combination treatment, which again exceeded either of the single agent treatments (Fig. 6B and Supplementary Fig. 10F).
A Cell viability of A549 cells treated with CsA (8 μM) and CB-839 (0.08 μM) combination (Com) in presence of 2, 1, 0.5 mM glutamine (Gln). B 3D clonogenic assay of A549-mCherry cells treated with vehicle, CsA, CB-839, or their combination (n = 3 independent experiments). After growing for 2 weeks, formed clones were quantitated by using Image J. The synergistic anti-proliferative effect was evaluated by Combenefit. Blue indicates synergy, while red indicates antagonism between drugs. C Lung orthotopic model was established by tail-vein injection of A549-Luc cells. Mice were divided into four groups (n = 6 mice per group): vehicle control (p.o. twice daily), CB-839 (150 mg/kg, p.o. twice daily), CsA (20 mg/kg, i.p. every 3 days), and CB-839/CsA combination (150 mg/kg, p.o. twice daily; 20 mg/kg, i.p. every 3 days). Bioluminescence imaging was performed every 7 days using a PerkinElmer IVIS Spectrum CT system. D Statistics of lung metastatic nodules in the lung orthotopic model with different treatments (n = 6 mice per group). E Kaplan–Meier survival analysis of different groups in lung orthotopic model formed by tail-vein injection of A549-luc cells (n = 6 mice per group). F, G Tumor growth in NCG mice bearing MT−101 F or WT-201 G PDX xenograft treated with the vehicle (p.o. twice daily), CB-839 (150 mg/kg, p.o. twice daily), CsA (20 mg/kg, i.p. every 3 days), and CB-839/CsA combination (n = 6 mice per group). MT−101 PDX harbors concurrent KEAP1 and KRAS mutations (KEAP1G333C and KRASG12D). WT-201 PDX has KEAP1WT and KRASWT. A represents mean ± SD of three independent experiments. P values were analyzed using Two-tailed unpaired Student’s t-test for A and D, using Log-rank (Mantel-Cox) test for E, and using One-way ANOVA for (F, G). P < 0.05 was considered statistically significant. Source data are provided as a Source Data file.
We next assessed the potency of combined CsA and CB-839 in vivo. Luciferase-labeled A549 (A549-Luc) cells were injected into the tail vein of nude mice, and tumor progression was monitored using an IVIS imaging system. Mice were administered with CsA (20 mg/kg, i.p., every three days), CB-839 (150 mg/kg, p.o., twice daily), or their combination. 3D bioluminescence imaging showed that the vehicle control animals developed massive tumors in their lungs. While both the CsA and CB-839 monotherapies reduced the tumor burden compared to the vehicle control, it was obvious that the combination therapy conferred superior anti-tumor effects (Fig. 6C). Consistently, analysis of isolated lung tissues also showed that the CsA and CB-839 combination therapy resulted in significantly fewer tumor foci compared to the vehicle control and compared to both monotherapies (Fig. 6D). It was also highly notable that the CsA and CB-839 drug combination significantly extended the survival of tumor-bearing animals (Fig. 6E; P < 0.0001). Note that there was no obvious change in mouse body weight in any of the groups, suggesting that these treatments were well tolerated by the animals (Supplementary Fig. 11A). Moreover, considering the potential immunosuppressive activity of CsA, we also examined the efficacy of CsA and CB-839 drug combination using humanized NCG mice with A549-implanted tumors. Of note, CsA and CB-839 combination therapy have similar anti-tumor activity in immune-competent humanized NCG mice, compared with immunodeficient nude mice (Supplementary Fig. 11B).
Finally, the in vivo efficacy of CsA and CB-839 drug combination was evaluated in clinically relevant patient-derived xenograft (PDX) models. We employed five NSCLC PDX models with distinct genotypes: the MT-101 and MT-102 models harbor concurrent mutant KEAP1/KRAS, while the WT-201, WT-202 and WT-203 models exhibit KEAP1/KRAS WT (Supplementary Table 3). Consistent with the co-occurring KEAP1/KRAS genotype, we detected an obvious increase in the NRF2 protein level in the MT-101 tumors compared to WT-201 tumors (Supplementary Fig. 11C). For the MT-101 PDX model, the CsA and CB-839 monotherapies each moderately significantly reduced the tumor volumes, with inhibition rates of 29.83 ± 9.50% and 49.41 ± 5.91%, respectively, compared to the vehicle control. The CsA and CB-839 combination therapy performed better, achieving an inhibition rate of 75.17 ± 4.20% (Fig. 6F and Supplementary Fig. 11D). We also examined the anti-tumor synergy of CsA and CB-839 in MT-101 PDX model using a Q value method33,34,35. The Q value of CsA plus CB-839 was 1.165 (Q ≥ 1.15 indicates synergism), suggesting a synergistic effect between CB-839 and CsA. Of note, we observed a similar anti-tumor potency of CsA and CB-839 combination therapy in MT-102 PDX model (Supplementary Fig. 11F). By contrast, for KEAP1/KRAS WT PDX models we detected no significant differences in tumor growth between the vehicle control and any of the three treatments (CsA, CB-839, or their combination) (Fig. 6G, Supplementary Fig. 11E, G and H). These findings support that the observed therapeutic benefits of the CsA and CB-839 combination can be attributed to NRF2 hyperactivation.
Consistent with our earlier observations about the NRF2/KLF5/SLC1A5 pathway in cultured cells, immunoblotting of the MT-101 tumor tissues showed that the accumulation of NRF2, KLF5, and SLC1A5 was obviously reduced upon CsA treatment and upon the CsA and CB-839 combination treatment (Supplementary Fig. 11I). In contrast, CsA alone or combined with CB-839 have minor effects on NRF2, KLF5, and SLC1A5 proteins derived from WT-201 tumor samples (Supplementary Fig. 11J). Collectively, these results demonstrate that simultaneously targeting of PPIA and glutaminase represents a promising strategy for the treatment of NSCLC patients harboring NRF2 hyperactivation.
Elevated PPIA and NRF2 accumulation is positively associated with poor prognosis in NSCLC patients
To explore the clinical significance of PPIA and NRF2 in NSCLC, we examined their accumulation using a tumor tissue microarray comprising specimens of multiple diagnosed clinical stages. According to the immunohistochemical staining of paired tumor/normal tissues, PPIA and NRF2 expression were dramatically higher in NSCLC specimens than in adjacent normal lung tissues (Fig. 7A). Moreover, approximately 86.48% of the PPIA-high samples showed high expression level of NRF2, whereas 65.62% of clinical NSCLC specimens with low level of PPIA exhibited low NRF2 expression (Table 3, P < 0.001). Quantitation of the immunohistochemical staining also showed highly correlation of PPIA and NRF2 in NSCLC tissues (Fig. 7B, Pearson R = 0.6996, P < 0.0001). Further Kaplan-Meier survival analysis revealed that patients with high PPIA or high NRF2 expression had poorer prognosis than those with low PPIA or low NRF2 expression (Fig. 7C, D; Logrank P < 0.0001 for PPIA and Logrank P = 0.0117 for NRF2, respectively).
A Representative immunohistochemical analysis of PPIA and NRF2 in human LUAD tissue microarray containing lung tumor tissues and adjacent normal lung tissues. Scale bar = 50 μm. B Scatter plots showing a positive correlation of PPIA and NRF2 expression in IHC analysis of human NSCLC cancer tissues (n = 69 samples). Linear regression with Pearson R and two-tailed P values are shown. C Kaplan-Meier survival curves of patients with NSCLC divided by high or low PPIA protein expression level according to IHC analysis (n = 69 samples). D Kaplan-Meier survival curves of patients with NSCLC divided by high or low NRF2 protein expression level according to IHC analysis (n = 69 samples). E–H Kaplan–Meier survival curves of patients with NSCLC based on PPIA, NRF2, KLF5 and SLC1A5 gene expression level (for PPIA, n = 719 samples; for NRF2, n = 672 samples; for KLF5, n = 719 samples; for SLC1A5, n = 719 samples). Data are integrated from Kaplan–Meier plotter (http://kmplot.com/analysis/). Statistical significance for Kaplan–Meier survival curves (C–H) was calculated by Log-rank (Mantel-Cox) test. Source data are provided as a Source Data file.
We also analyzed the prognostic significance of PPIA and NRF2 using the Kaplan-Meier plotter database36. In line with the results of the tissue microarray, high mRNA expression of PPIA and NRF2 were associated with reduced overall survival time as well (Fig. 7E-7F; Logrank P < 0.0001 for PPIA and Logrank P < 0.0001 for NRF2, respectively). Additionally, we noted that NSCLC patients with higher KLF5 and SLC1A5 mRNA expression predict poor prognosis (Fig. 7G-7H; Logrank P = 0.0017 for KLF5 and Logrank P < 0.0001 for SLC1A5, respectively). Together, these data demonstrate that PPIA and NRF2 are positively associated with malignant progression and poor prognosis of NSCLC.
Discussion
More than 5% of the cancer patients were diagnosed as NRF2-mutant/hyperactivated cancers, as revealed by TCGA Pan-Cancer analysis, with significant enrichment in lung malignancy37,38,39. Accumulating evidence has established that NRF2 hyperactivation as a driver of cancer progression, metastasis, and therapy resistance4. Although the therapeutic appeal of NRF2 inhibition, the transcription factor is a tricky target. Research efforts have focused on alternative strategies aimed at exploring NRF2-related vulnerabilities. In the current study, we demonstrate that PPIA is essential for NRF2 stability and is a druggable vulnerability in NRF2-hyperactivated NSCLC.
NRF2 is a fundamental regulator of homeostatic milieu in response to diverse stress stimuli in most metazoans40,41. NRF2 protein in the cell was tightly controlled by ubiquitin proteasomal pathway42. It has previously been reported that NRF2-ECH homology (Neh) domains, such as Neh2, are critical determinants of NRF2 protein stability43. Herein, we demonstrated the existence of a specific primate-conserved sequence PBM located in the linker between the Neh4 and Neh5 domains, which is indispensable for governing NRF2 protein stability. PPIA binds to the P174-containing PBM fragment of NRF2 and blocks the access of ubiquitin/KEAP1 to NRF2, thus inhibiting ubiquitin-proteasome dependent degradation. P174A mutant fails to bind to PPIA with a shorten half-life. Our high-resolution co-crystal structure reveals that the trans P174-containing hydrophobic fragment is embedded in the catalytic pocket of PPIA. These findings open a new horizon of the complex molecular mechanisms that govern NRF2 protein stability in health and disease. Intriguingly, TCGA analysis indicated that the interaction interface of PPIA and NRF2 does not harbor mutations in lung and other cancers (Supplementary Fig. 12A, B). The biochemical and structural insights into the regulation of transcription factor NRF2 by PPIA may offer new therapeutic approaches for clinical interventions through disturbing the protein-protein interaction44.
PPIA exhibits peptidylprolyl cis-trans isomerase activity, which modulates substrate protein folding/stability and cell signaling45. PPIA has been identified as a dysfunctional protein in a variety of human cancers46,47,48. PPIA promotes the colonization and proliferation of multiple myeloma cells via binding to CD14749. A recent study further identified PPIA as an attractive target for the treatment of resistant multiple myeloma50. Charalampos G Kalodimos group have reported that PPIA binds to and enhances activation of CrkII, which stimulate breast cancer cell migration51. Yet, the molecular mechanisms underlying PPIA action and the signaling pathways in NSCLC are still poorly understood. In this study, we discovered that PPIA modulates NRF2 stability, and its prolyl cis-trans isomerase activity renders cancer progression of NRF2-hyperactivated NSCLC. However, we cannot completely exclude the implication of other PPIA substrate proteins for PPIA’s oncogenic properties in NSCLC cells, despite we found that two known substrate proteins CrkII and TARDBP are not affected by PPIA inhibition/KO (Supplementary Fig. 12C, D). Of note, tumor tissue microarray and bioinformatics analysis indicated that PPIA and NRF2 are vastly upregulated in patients with NSCLC. Our findings highlight a potential mechanism through which PPIA regulate NRF2 stability and reveal an appealing translational value of targeting PPIA in NRF2-hyperavtivated malignancies. In addition, PPIA is known to regulate transcription factor NFAT, which plays important roles in cancer progression52. Our findings raise the possibility that PPIA may act as a molecule node for the cross-talk between NRF2 and NFAT signaling in cancer. Further molecular mechanistic understanding of the reciprocal interactions between NRF2 and NFAT pathway may inspire novel biological insights and therapeutic regimens aimed at targeting tumor progression.
The FDA-approved macrocyclic peptide CsA impairs the binding of NRF2 to PPIA and triggers NRF2 degradation via ubiquitin-proteasome system. CsA represents a chemical inducer of NRF2 degradation. As a result, NRF2-hyperactivated NSCLC cell lines harboring KEAP1 and/or KRAS mutations, are highly sensitive to CsA treatment. And CsA significantly impairs the growth of NRF2-hyperactivated tumors in the clinically relevant PDX models. CsA is a widely used immunomodulatory drug, included in the World Health Organization (WHO) Model List of Essential Medicines (https://list.essentialmeds.org/?query=ciclosporin), with well-documented effectiveness and safety profile53,54. Clinically, CsA has been investigated for the possibility of being repurposed for the treatment of various cancers (https://www.clinicaltrials.gov). Our study raises the possibility that a deeper understanding of CsA’s mechanism will inspire a more reasonable therapeutic regimen in future clinical investigations using new, optimized PPIA inhibitors to treat patients with NRF2-hyperactivated NSCLC.
During cancer progression, NRF2 signaling functions as a master mechanism in response to hostile metabolic microenvironment55. Several pioneering works have established that NRF2 activation in cancer cells leads to increased glutamine dependence via enhancing consumption of glutamate for fueling tricarboxylic acid cycle, glutathione synthesis and glutamate excretion56,57,58. To sustain intracellular glutamate content, NRF2-hyperactivated cells use glutamine transporter SLC1A5 to uptake greater extracellular glutamine, which is subsequently converted to glutamate by glutaminase59. The high requirement of glutamine in NRF2-hyperactivated cells indicates a potential dependency of NRF2-hyperactivated cells on SLC1A532,60. SLC1A5 is up-regulated in the NRF2-hyperactivated cells, however, the underlying regulatory mechanisms of SLC1A5 expression by NRF2 are not fully understood. In this study, we demonstrated that oncogenic transcription factor KLF5, acting as a NRF2 target gene, activates the transcription of SLC1A5 via a specific GC-rich motif sequence. While, given the pleiotropic roles of NRF2 activation, it will be intriguing to investigate whether other transcriptional or posttranslational mechanisms contribute to NRF2-mediated SLC1A5 augmentation.
Recent studies have evidenced that KRAS-driven lung and pancreatic cancer with KEAP1 or NRF2 mutations are highly dependent on glutaminolysis and are vulnerable to glutaminase inhibition32,61. CB-839, a first-in-class glutaminase inhibitor, is actively explored as metabolic intervention for the treatment of multiple types of cancer, but its clinical benefit is limited as monotherapy62. Several phase I studies have demonstrated encouraging results for CB-839 as part of a combination regimen63. However, a recent phase II study combining CB-839 with pembrolizumab/chemotherapy as first-line treatment for KEAP1/NRF2-mutated metastatic NSCLC have been terminated due to lack of efficacy (NCT04265534)64. Despite these unsatisfactory outcomes, careful development and rational design of combination therapies with CB-839 might confer clinical benefits. Here, we have explored a combination therapy of CsA and CB-839. In both cell line-derived and patient-derived xenograft models with NRF2 hyperactivation, this combination treatment showed encouraging anti-cancer activity and tolerability. Our results may provide a rationale for exploiting CsA to sensitize patients with NRF2-hyperactivated tumors to glutaminase inhibition. However, extensive clinical investigations are required to optimize safe and effective combination therapy regimens of CsA and CB-839 to reduce drug-drug interaction and avoid drug resistance.
In summary, our study provides actionable mechanistic insights about NRF2 stability and reveals PPIA as a promising target for treating NRF2-hyperactivated lung cancer. Of note, the FDA-approved CsA, as a chemical inducer of NRF2 degradation, exhibits a significantly synergistic anti-cancer effect when administered as a combination therapy alongside the clinical glutaminase inhibitor CB-839 in NRF2-hyperactivated NSCLC. Our findings provide a potential framework toward precision medicine of this refractory cancer type in clinical trials.
Methods
Ethical statement
The experimental protocols of animal studies were approved by Institutional Animal Care and Use Committee (IACUC) of East China University of Science and Technology (ECUST-2021-10001).
Cell cultures and reagents
HEK293T cells and lung cancer cell lines, including A549, H2030, H1299, H1395, H1650, H460, H1944, H1703, H1975, H2122, H23, H292, H358 and H838 were kindly provided by Stem Cell Bank, Chinese Academy of Sciences. EBC-1 was purchased from Shanghai Hongshun Biotechnology Co., Ltd. H441 and H522 cell lines were purchased from ATCC (Manassas, VA, USA). All the cell lines have been authenticated by STR (short tandem repeat) profiling and confirmed mycoplasma free by PCR-based assay. All cell lines were cultured in DMEM medium supplemented with 10% FBS, 100 unit/mL penicillin and 100 µg/mL streptomycin and kept in a 37 °C humidified incubator containing 5% CO2.
The detailed information of reagents used in this study were provided in Supplementary Data 2. Antibody dilutions: NRF2 (Proteintech, 16396-1-AP; 1:1000 for WB; 2 μg for IP; 2 μg for ChIP; 1:100 for IHC), Histone-H3 (Proteintech, 17168-1-AP; 1:1000 for WB), PPIA (Proteintech, 10720-1-AP; 1:1000 for WB; 1:100 for IHC), Ubiquitin (Santa Cruz, sc-8017; 1:1000 for WB), SLC1A5 (Proteintech, 20350-1-AP; 1:1000 for WB), KLF5 (Proteintech, 21017-1-AP; 1:1000 for WB; 2 μg for ChIP), KEAP1 (Proteintech, 10503-2-AP; 1:1000 for WB), CRK (Proteintech, 16685-1-AP; 1:1000 for WB), TDP-43 (Proteintech, 10782-2-AP; 1:1000 for WB), HA (Proteintech, 51064-2-AP; 1:1000 for WB), Flag (Sigma-Aldrich, F3165; 1:1000 for WB; 2 μg for IP), GAPDH (Proteintech, 60004-1-Ig; 1:3000 for WB), IgG heavy chain (Proteintech, 16402-1-AP; 2 μg for IP).
RIG-NRF2 Screen
To assess NRF2 protein degradation, we developed a fluorescence probe named RIG-NRF2 (DsRed-IRES-EGFP-NRF2). Briefly, NRF2 was fused with green fluorescent protein EGFP and co-expressed with red fluorescent protein DsRed within a single transcription unit, which was separated by an internal ribosome entry site (IRES) motif. The cDNA of human NRF2 was amplified through polymerase chain reaction (PCR) using followed cloning primers:
FW: 5’-GATAATATGGCCACAATCGATATGATGGACTTGGAG-3’
RV: 5’-CACCATAGCGGCCGCCTCGAGGTTTTTCTTAACATC-3’.
Subsequently, the PCR product was cloned into the bicistronic lentiviral vector pLenti-DsRed_IRES_MAPT:EGFP vector (Addgene, Plasmid #92196) using one step cloning method.
HEK293T cells were transfected with RIG-NRF2 as well as packaging plasmids psPAX2 (Addgene plasmid #12260) and pMD2.G (Addgene plasmid #12259) using Lipofectamine 2000 reagent to generate lentivirus particles. A549 cells were then infected with lentivirus and selected with G418. The generated A549 subclone stably expressing RIG-NRF2 was named as A549-RIG-NRF2. To detect NRF2 protein level, A549-RIG-NRF2 cells were seeded into 6-well plates at the confluence of 70%-80% and then treated with MG132, cycloheximide (CHX), Ki696, ML334 or compounds for indicated times. Then, treated cells were washed with PBS for 3 times and resuspended in PBS at the concentration of 1 ×106 cells/mL. Fluorescence signal was detected by BioTek neo2 multi-mode reader or flow cytometry (Beckman coulter Cytoflex LX).
Plasmid construction
NRF2-K7R plasmid was kindly provided by Donna D. Zhang (University of Arizona, Tucson, USA) and the PCR-generated DNA containing NRF2-K7R was cloned into pCDNA3.1-Flag-Nanoluc using ClonExpress II One Step Cloning Kit. Site-direct mutagenesis and fragment deletion of PPIA and NRF2 were performed using KOD-Plus-Mutagenesis Kit according to the manufacturer’s instructions. Promoter region of SLC1A5 (-2,000 to +100 bp) and promoter region of KLF5 (-4,000 to 0 bp) were cloned into KpnI and XhoI sites of pGL3-Basic using ClonExpress II One Step Cloning Kit.
Luciferase reporter assay
Antioxidant response element (ARE)-driven reporter gene expression was performed as described previously65. Briefly, cells were seeded into white 96-well plates (15,000 cells per well) and subsequently co-transfected with pGL4.37[luc2P/ARE/Hygro], pCDNA3.1-NRF2-Flag and pSV40 vector with a ratio of 2:1:0.5 (totally 100 ng DNA/well) using lipofectamine 2000 reagent (Invitrogen, Grand Island, NY, USA). After 4 h-transfection, cells were treated with different concentrations of CsA for another 48 h. Luciferase activity was measured using a Dual-Glo® Luciferase Assay System (Promega, Madison, WI, USA) according to the manufacturer’s instructions.
Promoter activity assay
HEK293T cells were co-transfected with KLF5 promoter luciferase construct and pCDNA3.1-Flag-NRF2 or SLC1A5 promoter luciferase construct and pCDNA3.1-Flag-KLF5 using lipofectamine 2000. The transfected cells were seeded into 96-well plates and allowed to grow for 36 h. Subsequently, cells were lysed and the luciferase activities were detected by Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) using BioTek neo2 multi-mode reader (BioTek, Winooski, VT, USA).
CRISPR/Cas9 mediated PPIA gene knockout (PPIA KO)
PPIA gene knockout cancer cells were generated by using CRISPR/Cas9-mediated genome editing66. The lentiCRISPRv2 (Addgene, #52961) containing the specific sgRNA targeting PPIA gene and the packaging plasmid psPAX2 (Addgene plasmid #12260) and pMD2.G (Addgene plasmid #12259) was co-transfected into HEK293T cells. Two distinct sgRNAs targeting human PPIA gene were designed as following: sgPPIA#1: TTCTTCGACATTGCCGTCGA, and sgPPIA#2: ACAAGGTCCCAAAGACAGC. Lentivirus infection of NSCLC cells was conducted in the presence of polybrene (8 μg/mL) for 12 h. PPIA-KO cells were selected using puromycin (5 μg/mL) after expansion for 72 h and re-plating at high dilution in the presence of 5 μg/mL of puromycin in order to obtain individual colonies. Ablation of PPIA was verified using immunoblot.
Immunoblot analysis
Cells were washed with cold PBS twice and lysed in lysis buffer (supplemented with 1% protease inhibitor cocktail) at 4 °C. Protein from lysed cells was resolved by SDS-PAGE and transferred to PVDF membranes. After being blocked in 5% bovine serum albumin in TBST (120 mM Tris, 150 mM NaCl, 0.05% Tween20, pH=7.4) at room temperature for 1 h, the membranes were incubated with different primary antibodies overnight at 4 °C. Subsequently, the membranes were washed with TBST for 3 times and incubated with HRP-conjugated secondary antibodies at room temperature for 3 h. After washing with TBST for 4 times, the protein bands were detected by ECL Western Blotting Substrate Kit (Thermo Scientific, MA, US).
Ubiquitination assay
Cells were pre-treated with MG-132 (10 μM) for 4 h and lysed in lysis buffer (50 mM Tris, 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate, 1 mM EDTA pH=7.4, supplemented with 1% protease inhibitor cocktail). Cell lysates were subjected to immunoprecipitation using primary anti-NRF2 antibody at 4 °C overnight. The immunoprecipitated proteins were eluted by boiling with SDS-loading buffer at 100 °C for 5 min and detected by immunoblot analysis using specific antibody against Ubiquitin.
NanoBRET Assay67
HEK293T cells were seeded into 96-well clear-bottom white plates (20,000 cells per well) and then co-transfected with pCDNA3.1-NRF2-Nanoluc and HaloTag-Ubiqiuitin with a ratio of 1:100 (totally 300 ng DNA/well) using lipofectamine 2000 reagent (Invitrogen, Grand Island, NY, USA). 24-h after transfection, cells were incubated with 100 nM HaloTag NanoBRET 618 Ligand (Promega, Madison, WI, USA) for 24 h at 37 °C. BRET signals were detected by adding Nanoluc substrate furimazine (10 μM) with BioTek neo2 multi-mode reader (BioTek, Winooski, VT, USA).
Quantitative PCR (Q-PCR)
Total RNA of cells or tumor tissues was extracted with GenEluteTM Total RNA purification Kit and cDNA was prepared using Hifair® II 1st Strand cDNA Synthesis SuperMix (Yeasen, Shanghai, China). Quantitative reverse-transcription PCR was conducted using Hifair® III One Step RT-qPCR SYBR Green Kit (Yeasen, Shanghai, China) according to its manufacturer’s instructions. The fold change of genes was calculated by 2-ΔΔCT method taking GAPDH as housekeeping gene (ΔCt = ΔCttarget - ΔCtGAPDH; ΔΔCt = ΔCtsample -ΔCtcontrol) and normalized with control groups which were defined as 1.0. All primers sequences were listed in Supplementary Data 1.
Cell viability assay
Cell viability was determined by using MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) method as described previously68. In brief, cells were seeded into 96-well plates and then treated with indicated concentrations of compounds for 96 h. Subsequently, cells were incubated with MTT reagent with a final concentration of 0.5 mg/mL for 4 h at 37 °C. The generated formazan crystals were dissolved in 150 μL dimethyl sulfoxide (DMSO) and the optical density at 570 nm was examined using BioTek neo2 multi-mode reader (BioTek, Winooski, VT, USA). The combination index (CI) of drug combinations was calculated using CalcuSyn software as described previously68. CI values < 1, =1, and >1 indicate synergism, additive, and antagonism, respectively.
3D colony formation assay
For 3D colony formation assay69, 0.7% and 1.2% BactoTM agar solution (BD Biosciences, San Jose, CA) were prepared, respectively. The mixture of 2x cell culture medium and 1.2% agar (in a 1:1 ratio) was seeded into 6-well plates as the bottom layer and the mixture of 2x cell culture medium containing NSCLC cancer cells stably expressing mCherry and 0.7% agar (in a 1:1 ratio) was plated as upper layer. Fresh culture medium containing various concentrations of compounds was maintained over the upper layer and changed every 3 days. After growing for 2 weeks, colonies were imaged using the fluorescence microscope (Olympus, Japan).
Co-immunoprecipitation (Co-IP) assay
HEK293T cells were transiently transfected with different expression vectors. After 36 h, cells were washed with cold PBS and lysed with lysis buffer (50 mM Tris pH 8.0, 150 mM NaCl, 10% glycerol, 1 mM EDTA, 50 mM NaF, and 0.1% NP-40) supplemented with 1% protease inhibitor cocktail. Endogenous proteins derived from NSCLC cells were also used for co-immunoprecipitation. The whole cell lysates were incubated with specific antibody and protein A/G Sepharose beads overnight at 4 °C. Subsequently, the immunocomplexes were washed with lysis buffer for 3 times at 4 °C and eluted with loading buffer at 100 °C for 5 min. Protein sample was resolved by SDS-PAGE and immunoblot.
Oxygen consumption rate (OCR) analysis
The OCR was analyzed using Seahorse XFe96 Analyzer (Agilent Technologies) according to the manufacturer’s instruction70. A total of 1.5 × 104 cells were seeded per well into 96-well cell culture plates (Agilent Technologies) and cultured with 10% FBS supplemented DMEM overnight at 37 °C incubator containing 5% CO2. Next day, the medium was removed and replaced with XF DMEM Base Medium, pH=7.4 (Agilent Technologies) supplemented with 2 mM glutamine (Agilent Technologies) and the cells were incubated in 37 °C non-CO2 incubator to balance the CO2 level. Using the XFe96 Analyzer, OCR was measured in baseline conditions (basal OCR) and metabolic stress conditions (maximal OCR) induced by several metabolic drugs, including oligomycin (1 μM), FCCP (0.5 μM), and Rotenone/Antimycin (0.5/0.5 μM) using Seahorse XF Cell Mito Stress Test kit (Agilent Technologies). Each measurement consisted of 3 cycles: 3 min Mix, 0 min Wait and 3 min Measure. The OCR values were normalized according to cell number and calculated with WAVE software (Agilent Technologies).
Chromatin immunoprecipitation (ChIP)
ChIP assay was performed using ChIP Assay Kit (Beyotime Biotechnology, Shanghai, China) according to the manufacturer’s protocol71. 5×106 cells were harvested and fixed with 1% formaldehyde for 10 min at 37 °C. The reaction was quenched by 125 mM glycine for 10 min at room temperature. The DNA of cell nuclei was isolated and sheared to 200-500 bp through sonication. Fragmented chromatin was subjected to immunoprecipitation reactions using Protein A/G agarose beads and anti-NRF2 (5 μg), anti-KLF5 (5 μg) or control IgG (5 μg) at 4 °C. The beads were then washed with Low-Salt Immune Complex Wash Buffer, High-Salt Immune Complex Wash Buffer, LiCl Immune Complex Wash Buffer and TE Buffer (twice) for 3 min at 4 °C. DNA-protein complexes were de-crosslinked by treating with 0.2 M NaCl and heating (65 °C, 4 hours). The proteins were removed by protein K treatment. The resultant DNA product was subjected to PCR analysis. The human KLF5 or SLC1A5 promoter-specific primers sequences were shown in Supplementary Data 1.
siRNA transfection
siRNA negative control and siRNA targeting NRF2, PPIA, KLF5 were obtained from Shanghai Synbio Technologies. For siRNA transfection, cells were seeded at about 60% confluence into 6 cm dishes and then transfected with specific siRNA using lipofectamine 3000 reagent following the manufacturer’s manual for 36 h. The gene knockdown efficiency was confirmed by immunoblot. The siRNA sequences were listed in Supplementary Data 1.
Recombinant protein expression and purification
Recombinant PPIA protein was expressed in BL21 (DE3) E. coli after induction with 0.2 mM IPTG (isopropyl-β-D-thiogalactopyranoside) overnight at 25 °C72. 15N-labeled PPIA protein was produced from E. coli grown at 16 °C. Cell pellets were collected and lysed by sonication in lysis buffer (25 mM Tris, 300 mM NaCl, 5% glycerol, pH=8.0). Then, the clarified lysate was loaded onto the Ni-NTA column (Thermofisher, MA, US) and washed with lysis buffer containing 20 mM imidazole. The bound target protein was eluted with lysis buffer containing 200 mM imidazole.
Pull-down assay
Purified PPIA protein was conjugated with CNBr-activated Sepharose 4B beads (GE Healthcare, UK) according to standard manufacturer’s protocol72. Firstly, the CNBr-activated Sepharose 4B beads were suspended and washed in 1 mM HCl and then washed with coupling buffer, 0.1 M NaHCO3 pH=8.3 containing 0.5 M NaCl. Secondly, activated beads were incubated with or without purified PPIA protein in coupling buffer overnight at 4 °C and subsequently washed with coupling buffer and blocked with 0.1 M Tris pH=8.0 for 2 h. Thirdly, protein-conjugated beads were washed five cycles of different pH washing buffers (buffer 1, 0.1 M acetate, 0.5 M NaCl, pH 4.0; buffer 2, 0.1 M Tris, 0.5 M NaCl, pH 8.0) and incubated with cell lysate in binding buffer (25 mM HEPES, 0.15 M NaCl, 1 mM DTT, pH=7.5) for 3 h at 4 °C. Lastly, the beads were washed with binding buffer for 5 times and eluted with SDS gel-loading buffer for 5 min at 100 °C followed by immunoblot analysis.
Glutamine assay
Treated cells were plated at 20,000 cells/well in 96-well plates in DMEM medium supplemented with 4 mM glutamine and 10% dialyzed FBS. Cellular glutamine level was measured using the glutamine/glutamate-Glo Assay kit (Promega, Madison, WI, USA) following manufacturer’s instruction.
Stable-isotope labeling and metabolic flux analysis73
For 13C5-glutamine labeling, treated cells (2 × 106) were cultured in glutamine free DMEM medium containing 10% dialyzed serum and 2 mM 13C5-glutamine for 8 h. After labeling, cells were washed twice with 37 °C pre-warmed saline, lysed by 1 mL of 80% ice-cold methanol solution, and sonicated five times for 5 s, with 5 s intervals between treatments. The samples were centrifuged at 18,000 g for 10 min at 4 °C and all the supernatant were collected and stored at -80 °C until quantitative analysis.
Metabolites were quantitated by Metabo Profile Biotechnology (Shanghai, China). Samples were thawed on an ice bath to reduce sample degradation before processing. 400 μL 80% methanol solution was added to cell samples. Then, the samples were sonicated and centrifuged at 18,000 g for 15 minutes at 4 °C. All the supernatant after centrifugation was collected for LC-MS analysis.
For LC-MS analysis, the samples were analyzed by ultra-performance liquid chromatography coupled to tandem mass spectrometry (UPLC-MS/MS) system (ACQUITY UPLC-Xevo TQ-S, Waters Corp., Milford, MA, USA). The optimized instrument settings are briefly described as follows. For HPLC, column: UPLC HILIC (4.6-mm column, Amide XBridge, Waters), column temperature: 40 °C, sample manager temperature: 10 °C, mobile phases: A = 5% acetonitrile in water (with 20 mM ammonium hydroxide and ammonium acetate); and B = acetonitrile, gradient conditions: 0-3.5 min (85-32% B), 3.5-12 min (32-2% B), 12-16.5 min (2% B), 16.5-17 min (2-85% B), 17-25.5 min (85% B), flow rate: 0.40 mL/min, and injection vol.: 5.0 µL. For mass spectrometer, capillary 2.5 (ESI-) Kv, source temperature: 150 °C, desolvation temperature: 550 °C, and desolvation gas flow: 1000 L/h. The raw data files generated by UPLC-MS/MS were processed using the MassLynx software (v4.1, Waters, Milford, MA, USA) to perform peak integration, calibration, and quantitation for each metabolite.
Nuclear magnetic resonance spectroscopy
[1H, 15N] HSQC (heteronuclear single quantum coherence) experiments of 15N-labeled PPIA were performed in the presence or absence of 5-fold molar excess of NRF2 fragment PBM 169VAQVAPVD176 at 25 °C on a Bruker Avance III 600 MHz NMR spectrometer equipped with a cryogenically cooled probe. All of the NMR samples were prepared using buffer containing 100 mM NaCl, 50 mM Bis-Tris, pH 7.4 with 10% D2O, and the final concentration of PPIA was 500 μM. NMR data processing and analysis were performed by using the programs of NMRPipe1 and Sparky (Goddard and Kneller, Sparky 3, University of California, San Francisco). The amide resonance assignments of PPIA extracted from the BMRB entry of 27620 were used in the chemical shift perturbation (CSP) analysis, and the CSP values (Δδ) for 15N and 1H nuclei were calculated according to Eq. 1, where ΔδN and ΔδH represent the CSP values of the amide nitrogen and proton, respectively.
Crystallization and structure determination
For crystallization, the recombinant PPIA protein was further purified through gel filtration (Superdex 75, GE Healthcare) in buffer containing 25 mM Hepes, 100 mM NaCl, 0.5 mM TCEP with pH value of 7.5. The purified protein was concentrated into 50 mg/mL and incubated with indicated NRF2 fragment PBM (169VAQVAPVD176, 2 mM) at 4 °C for 2 h. Hanging-drop vapor-diffusion in 24-well crystallization plates (Hampton Research, USA) was used for co-crystallization. The PPIA/NRF2-PBM complex was mixed with equal volume of reservoir solution consisting of 25 mM Hepes, 25% PEG 3350, 5 mM TCEP with pH value of 7.5. The cubic complex crystal appeared within two weeks and then mounted in a 0.3 μm loop with addition cryoprotectant of 30% Tween 20 and flash-cooled in liquid nitrogen before data collection. X-ray diffraction data was collected at the synchrotron beamline BL19U1 in SSRF, Shanghai, China. The data was processed using HKL3000 program suite (HKL Research, Charlottesville, VA), phased by Molrep (CCP4) with PDB entry 3K0M as search model and further refined via Phenix as well as Coot. Data quality and processing statistics were summarized in Table 2 and Supplementary Table 2. The images of complex structure were prepared using PyMOL (Molecular Graphics System, Version 2.2.3; Schrödinger, LLC). The structure factors of the final complex crystal were deposited in the Protein Data Bank (PDB, www.rcsb.org, PDB ID: 8HZ8).
Mouse strains
Female BALB/c Nude mice were purchased from Shanghai Jiesijie Laboratory. Female NOD/ShiLtJGpt-Prkdcem26Cd52Il2rgem26Cd22/Gpt (NCG) mice and female CD34+ HSC-derived humanized NCG mice were purchased from Jiangsu GemPharmatech Company. Mice were housed in a dedicated animal facility designed for laboratory rodents. The temperature in the facility was maintained at a range of 20-24 °C with a relative humidity of 40-60%. A 12-hour light-dark cycle was maintained. All maintenance and experimental procedures involving animals were approved by the Institutional Animal Care and Use Committee of East China University of Science and Technology. The maximal tumor size permitted by the Institutional Animal Care and Use Committee was 3000 mm3.
Xenograft models in nude mice
A549 and H1650 cells with inducible shCtrl or shPPIA were injected subcutaneously at the back of 6-week-old female BALB/c nude mice (5×106 per mouse). The mice were given doxycycline (20 mg/kg) by oral gavage every other day when tumor volumes reached approximately 50 mm3. Tumor volumes were measured with calipers every 3 days till the control group volumes reached to around 500 mm3.
Xenograft models in humanized NCG mice
A549 cells (5×106 per mouse) were subcutaneously injected at the back of 18-week-old female CD34+ HSC-derived humanized NCG mice. When tumor volume reached to around 50 mm3, the mice were randomly divided into four groups: (1) Vehicle; (2) CB-839 (CB-839 in 25% (W/V) β-cyclodextrin pH=2, 150 mg/kg, p.o. twice daily); (3) CsA, (CsA in 10% DMSO + 10% Kolliphor EL + 80% PBS, 20 mg/kg i.p. every 3 days); (4) CB-839/CsA combination (CB-839 150 mg/kg, p.o. twice daily; CsA 20 mg/kg, i.p. every 3 days). Tumor volumes were measured with calipers every 3 days till the control group volumes reached to around 500 mm3.
Lung orthotopic model
Lung orthotopic model was established using luciferase-expressing A549 (A549-Luc) cells74. In brief, 6-week-old female BALB/c nude mice were injected with 100 μL A549-Luc cells (5×106 per mouse) through the lateral tail vein and lung lesion formation was monitored by bioluminescence imaging using D-luciferin (2 mg per mouse) every 7 days. Mice were randomly divided into four groups: (1) Vehicle; (2) CB-839 (CB-839 in 25% (W/V) β-cyclodextrin pH=2, 150 mg/kg, p.o. twice daily); (3) CsA, (CsA in 10% DMSO + 10% Kolliphor EL + 80% PBS, 20 mg/kg i.p. every 3 days); (4) CB-839/CsA combination (CB-839 150 mg/kg, p.o. twice daily; CsA 20 mg/kg, i.p. every 3 days). The body weight of each mouse was recorded weekly. At the end of the experiments, mice were euthanized and lung tissues were harvested, and fixed with bouin’s fluid. The number of lung nodules was recorded.
Patient-derived xenograft (PDX)
Human tumor specimens were supplied by Affiliated Tumor Hospital of Guangxi Medical University. Written informed consent was provided by participants and approved by Ethics Committee of Guangxi Medical University Cancer Hospital. The genomic DNA of tumor tissues derived from patients with lung adenocarcinoma was extracted using TIANamp Genomic DNA Kit (QIAGEN). Mutations in KRAS (exons 2 and 3) and KEAP1 (exons 2, 3, 4, 5) were analyzed using PCR amplification and DNA sequencing. Primers sequences were listed in Supplementary Data 1.
PDX study was performed as described previously with some modifications75. Briefly, tumor fragments were implanted at the back of 6-week-old female NCG mice. When tumor volumes reached approximately 1,000 mm3, tumor tissues were collected and re-implanted into NCG mice. When the third generation of tumors reached 150-200 mm3, those mice were randomly divided into 4 groups: (1) Vehicle (p.o. twice daily); (2) CB-839 (CB-839 in 25% (W/V) β-cyclodextrin pH=2, 150 mg/kg, p.o. twice daily); (3) CsA (CsA in 10% DMSO + 10% Kolliphor EL + 80% PBS, 20 mg/kg i.p. every 3 days); (4) CB-839/CsA combination (CB-839 150 mg/kg, p.o. twice daily; CsA 20 mg/kg, i.p. every 3 days). Mice were sacrificed when the vehicle group volumes reached to around 2000 mm3. Tumor volumes were measured with calipers every 3 days.
Tumor tissue microarray
Tissue microarray of human LUAD samples (n = 69) were obtained from Shanghai Superbiotek Pharmaceutical Technology Co. Ltd. (Shanghai, China). For immunohistochemical staining, the tissue microarray was dewaxed and rehydrated. Protein expression was detected by anti-PPIA or anti-NRF2 antibodies at 4 °C overnight and followed by HRP-labeled secondary antibodies. Subsequently, tissue microarray was probed by streptavidin-alkaline phosphatase system and scanned using 3DHISTECH (Hungary, PANNORAMIC Desk/MIDI/250/1000).
Bioinformatics analysis
The association of the gene expression level of KLF5 with that of SLC1A5 or NRF2 in NSCLC patients was analyzed in PrognoScan database (dataset GSE8894, http://www.prognoscan.org)27,76. To further examine the clinical significance of PPIA, NRF2, KLF5, and SLC1A5 expression, we assessed whether their expression levels were associated to survival benefits in NSCLC using Kaplan-Meier survival plot for grouping patients (Kaplan-Meier plotter, http://kmplot.com/analysis/)36. The risk differences were calculated by the log-rank test.
Statistics and reproducibility
Statistical analysis was conducted using GraphPad Prism 8.0 software (GraphPad Software, California, USA). Specially in Table 3, the correlation between NRF2 and PPIA expression was analyzed by bivariate Pearson correlation analysis using SPSS Statistics 28.0 software (IBM, Armonk, New York). Data were presented as mean ± S.D. for the specified number of independent experiments. Statistical significance between two groups were determined by unpaired two-tailed Student’s t-test. For Kaplan-Meier survival curves, Log-rank (Mantel-Cox) test was performed. For tumor growth inhibition, One-way ANOVA analysis was performed. The Pearson’s correlation method was utilized to perform the correlation test. P < 0.05 indicates statistically significant, P < 0.01 indicates very significant, P < 0.001 indicates highly significant. No data were excluded from the analyses.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The atomic coordinates and experimental data have been deposited in the Protein Data Bank (www.wwpdb.org) under accession code 8HZ8. The PPIA-CsA X-ray crystallographic data and PPIA-PBM search model used in this study are available in the Protein Data Bank (www.wwpdb.org) under accession code 1CWA and 3K0M. The publicly available NSCLC clinical data used in this study are available in the GEO database under accession code GSE889427. The remaining data associated with this study are present within the Article, Supplementary Information or Source Data file. Source data are provided with this paper.
References
Ma, Q. Role of nrf2 in oxidative stress and toxicity. Annu Rev. Pharm. Toxicol. 53, 401–426 (2013).
Jaramillo, M. C. & Zhang, D. D. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 27, 2179–2191 (2013).
Cuadrado, A. et al. Therapeutic targeting of the NRF2 and KEAP1 partnership in chronic diseases. Nat. Rev. Drug Discov. 18, 295–317 (2019).
Rojo de la Vega, M., Chapman, E. & Zhang, D. D. NRF2 and the hallmarks of Cancer. Cancer Cell 34, 21–43 (2018).
Menegon, S., Columbano, A. & Giordano, S. The dual roles of NRF2 in Cancer. Trends Mol. Med 22, 578–593 (2016).
Singh, A. et al. NRF2 Activation Promotes Aggressive Lung Cancer and Associates with Poor Clinical Outcomes. Clin. Cancer Res 27, 877–888 (2021).
DeNicola, G. M. et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature 475, 106–109 (2011).
Best, S. A. et al. Distinct initiating events underpin the immune and metabolic heterogeneity of KRAS-mutant lung adenocarcinoma. Nat. Commun. 10, 4190 (2019).
Lignitto, L. et al. Nrf2 activation promotes lung cancer metastasis by inhibiting the degradation of Bach1. Cell 178, 316–329.e318 (2019).
Bushweller, J. H. Targeting transcription factors in cancer - from undruggable to reality. Nat. Rev. Cancer 19, 611–624 (2019).
DeNicola, G. M. et al. NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat. Genet 47, 1475–1481 (2015).
Saigusa, D. et al. Impacts of NRF2 activation in non-small-cell lung cancer cell lines on extracellular metabolites. Cancer Sci. 111, 667–678 (2020).
Yen, H. C., Xu, Q., Chou, D. M., Zhao, Z. & Elledge, S. J. Global protein stability profiling in mammalian cells. Science 322, 918–923 (2008).
Blake, D. R. et al. Application of a MYC degradation screen identifies sensitivity to CDK9 inhibitors in KRAS-mutant pancreatic cancer. Sci. Signal 12, eaav7259 (2019).
Davies, T. G. et al. Monoacidic inhibitors of the kelch-like ech-associated protein 1: nuclear factor erythroid 2-related factor 2 (keap1:nrf2) protein-protein interaction with high cell potency identified by fragment-based discovery. J. Med Chem. 59, 3991–4006 (2016).
Hu, L. et al. Discovery of a small-molecule inhibitor and cellular probe of Keap1-Nrf2 protein-protein interaction. Bioorg. Med Chem. Lett. 23, 3039–3043 (2013).
Handschumacher, R. E., Harding, M. W., Rice, J., Drugge, R. J. & Speicher, D. W. Cyclophilin: a specific cytosolic binding protein for cyclosporin A. Science 226, 544–547 (1984).
Takahashi, N., Hayano, T. & Suzuki, M. Peptidyl-prolyl cis-trans isomerase is the cyclosporin A-binding protein cyclophilin. Nature 337, 473–475 (1989).
Zydowsky, L. D. et al. Active site mutants of human cyclophilin A separate peptidyl-prolyl isomerase activity from cyclosporin A binding and calcineurin inhibition. Protein Sci. 1, 1092–1099 (1992).
Zhang, D. D., Lo, S.-C., Cross, J. V., Templeton, D. J. & Hannink, M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol. Cell Biol. 24, 10941–10953 (2004).
Wu, T. et al. Hrd1 suppresses Nrf2-mediated cellular protection during liver cirrhosis. Genes Dev. 28, 708–722 (2014).
Baird, L. & Yamamoto, M. The molecular mechanisms regulating the KEAP1-NRF2 pathway. Mol. Cell Biol. 40, e00099–00020 (2020).
Mikol, V., Kallen, J., Pflügl, G. & Walkinshaw, M. D. X-ray structure of a monomeric cyclophilin A-cyclosporin A crystal complex at 2.1 A resolution. J. Mol. Biol. 234, 1119–1130 (1993).
Galan-Cobo, A. et al. LKB1 and KEAP1/NRF2 pathways cooperatively promote metabolic reprogramming with enhanced glutamine dependence in kras-mutant lung adenocarcinoma. Cancer Res 79, 3251–3267 (2019).
Mitsuishi, Y. et al. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell 22, 66–79 (2012).
Yang, C. et al. Glutamine oxidation maintains the TCA cycle and cell survival during impaired mitochondrial pyruvate transport. Mol. Cell 56, 414–424 (2014).
Lee, E.-S. et al. Prediction of recurrence-free survival in postoperative non-small cell lung cancer patients by using an integrated model of clinical information and gene expression. Clin. Cancer Res 14, 7397–7404 (2008).
Fornes, O. et al. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res 48, D87–D92 (2020).
Malhotra, D. et al. Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-Seq profiling and network analysis. Nucleic Acids Res 38, 5718–5734 (2010).
Stelzer, G. et al. The genecards suite: from gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinforma. 54, 1.30.31–31.30.33 (2016).
Stine, Z. E., Schug, Z. T., Salvino, J. M. & Dang, C. V. Targeting cancer metabolism in the era of precision oncology. Nat. Rev. Drug Discov. 21, 141–162 (2022).
Romero, R. et al. Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nat. Med 23, 1362–1368 (2017).
Jin, Z. About the evaluation of drug combination. Acta Pharm. Sin. 25, 146–147 (2004).
Duarte, D. & Vale, N. Evaluation of synergism in drug combinations and reference models for future orientations in oncology. Curr. Res Pharm. Drug Discov. 3, 100110 (2022).
Shen, N., Yang, C., Zhang, X., Tang, Z. & Chen, X. Cisplatin nanoparticles possess stronger anti-tumor synergy with PD1/PD-L1 inhibitors than the parental drug. Acta Biomater. 135, 543–555 (2021).
Lánczky, A. & Győrffy, B. Web-based survival analysis tool tailored for medical research (kmplot): development and implementation. J. Med Internet Res 23, e27633 (2021).
Cancer Genome Atlas Research, N. et al. The cancer genome atlas pan-cancer analysis project. Nat. Genet 45, 1113–1120 (2013).
Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012).
Cloer, E. W., Goldfarb, D., Schrank, T. P., Weissman, B. E. & Major, M. B. NRF2 activation in cancer: From DNA to protein. Cancer Res 79, 889–898 (2019).
Gacesa, R., Dunlap, W. C., Barlow, D. J., Laskowski, R. A. & Long, P. F. Rising levels of atmospheric oxygen and evolution of Nrf2. Sci. Rep. 6, 27740 (2016).
Kwak, M.-K. & Kensler, T. W. Targeting NRF2 signaling for cancer chemoprevention. Toxicol. Appl Pharm. 244, 66–76 (2010).
Sporn, M. B. & Liby, K. T. NRF2 and cancer: the good, the bad and the importance of context. Nat. Rev. Cancer 12, 564–571 (2012).
Poh, J. et al. A functionally defined high-density NRF2 interactome reveals new conditional regulators of ARE transactivation. Redox Biol. 37, 101686 (2020).
Cheng, F. et al. Comprehensive characterization of protein-protein interactions perturbed by disease mutations. Nat. Genet 53, 342–353 (2021).
Nigro, P., Pompilio, G. & Capogrossi, M. C. Cyclophilin A: a key player for human disease. Cell Death Dis. 4, e888 (2013).
Volker, S. E., Hedrick, S. E., Feeney, Y. B. & Clevenger, C. V. Cyclophilin a function in mammary epithelium impacts jak2/stat5 signaling, morphogenesis, differentiation, and tumorigenesis in the mammary gland. Cancer Res 78, 3877–3887 (2018).
Wang, G. et al. Cyclophilin a maintains glioma-initiating cell stemness by regulating wnt/β-catenin signaling. Clin. Cancer Res 23, 6640–6649 (2017).
Obchoei, S. et al. Cyclophilin A enhances cell proliferation and tumor growth of liver fluke-associated cholangiocarcinoma. Mol. Cancer 10, 102 (2011).
Zhu, D. et al. The Cyclophilin A-CD147 complex promotes the proliferation and homing of multiple myeloma cells. Nat. Med 21, 572–580 (2015).
Cohen, Y. C. et al. Identification of resistance pathways and therapeutic targets in relapsed multiple myeloma patients through single-cell sequencing. Nat. Med 27, 491–503 (2021).
Saleh, T. et al. Cyclophilin A promotes cell migration via the Abl-Crk signaling pathway. Nat. Chem. Biol. 12, 117–123 (2016).
Mancini, M. & Toker, A. NFAT proteins: emerging roles in cancer progression. Nat. Rev. Cancer 9, 810–820 (2009).
Landewé, R. B., van den Borne, B. E., Breedveld, F. C. & Dijkmans, B. A. Does cyclosporin A cause cancer? Nat. Med 5, 714 (1999).
Hijnen, D. J., ten Berge, O., Timmer-de Mik, L., Bruijnzeel-Koomen, C. A. F. M. & de Bruin-Weller, M. S. Efficacy and safety of long-term treatment with cyclosporin A for atopic dermatitis. J. Eur. Acad. Dermatol Venereol. 21, 85–89 (2007).
Hayes, J. D. & Ashford, M. L. J. Nrf2 orchestrates fuel partitioning for cell proliferation. Cell Metab. 16, 139–141 (2012).
Sayin, V. I. et al. Activation of the NRF2 antioxidant program generates an imbalance in central carbon metabolism in cancer. Elife 6, e28083 (2017).
LeBoeuf, S. E. et al. Activation of oxidative stress response in cancer generates a druggable dependency on exogenous non-essential amino acids. Cell Metab. 31, 339–350.e334 (2020).
Ding, H. et al. Activation of the NRF2 antioxidant program sensitizes tumors to G6PD inhibition. Sci. Adv. 7, eabk1023 (2021).
Okazaki, K., Papagiannakopoulos, T. & Motohashi, H. Metabolic features of cancer cells in NRF2 addiction status. Biophys. Rev. 12, 435–441 (2020).
Chan, K. et al. eIF4A supports an oncogenic translation program in pancreatic ductal adenocarcinoma. Nat. Commun. 10, 5151 (2019).
Mukhopadhyay, S. et al. Undermining glutaminolysis bolsters chemotherapy while nrf2 promotes chemoresistance in kras-driven pancreatic cancers. Cancer Res 80, 1630–1643 (2020).
Zhao, Y. et al. 5-fluorouracil enhances the antitumor activity of the glutaminase inhibitor cb-839 against pik3ca-mutant colorectal cancers. Cancer Res 80, 4815–4827 (2020).
Yang, W.-H., Qiu, Y., Stamatatos, O., Janowitz, T. & Lukey, M. J. Enhancing the efficacy of glutamine metabolism inhibitors in cancer therapy. Trends Cancer 7, 790–804 (2021).
Skoulidis, F. et al. A phase II randomized study of telaglenastat, a glutaminase (GLS) inhibitor, versus placebo, in combination with pembrolizumab (Pembro) and chemotherapy as first-line treatment for KEAP1/NRF2-mutated non-squamous metastatic non-small cell lung cancer (mNSCLC). J. Clin. Oncol. 38, TPS9627–TPS9627 (2020).
Ge, W. et al. iASPP is an antioxidative factor and drives cancer growth and drug resistance by competing with nrf2 for keap1 binding. Cancer Cell 32, 561–573.e566 (2017).
Sanjana, N. E., Shalem, O. & Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014).
Wang, E. et al. Modulation of RNA splicing enhances response to BCL2 inhibition in leukemia. Cancer Cell 41, 164–180.e168 (2023).
Wu, D. et al. Pharmacological inhibition of dihydroorotate dehydrogenase induces apoptosis and differentiation in acute myeloid leukemia cells. Haematologica 103, 1472–1483 (2018).
Liu, X. et al. A protein-fragment complementation assay reveals that celastrol and gambogic acid suppress ERα mutants in breast cancer. Biochem Pharm. 188, 114583 (2021).
Yoo, H. C. et al. A variant of slc1a5 is a mitochondrial glutamine transporter for metabolic reprogramming in cancer cells. Cell Metab. 31, 267–283.e212 (2020).
Liu, P. et al. RPA1 binding to NRF2 switches ARE-dependent transcriptional activation to ARE-NRE-dependent repression. Proc. Natl Acad. Sci. USA 115, e10352–e10361 (2018).
Lu, W. et al. Selective targeting p53(WT) lung cancer cells harboring homozygous p53 Arg72 by an inhibitor of CypA. Oncogene 36, 4719–4731 (2017).
Antoniewicz, M. R. A guide to 13C metabolic flux analysis for the cancer biologist. Exp. Mol. Med 50, 1–13 (2018).
Wang, X. et al. UDP-glucose accelerates SNAI1 mRNA decay and impairs lung cancer metastasis. Nature 571, 127–131 (2019).
Jiang, Y. et al. Establishment of lung cancer patient-derived xenograft models and primary cell lines for lung cancer study. J. Transl. Med 16, 138 (2018).
Mizuno, H., Kitada, K., Nakai, K. & Sarai, A. PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med Genomics 2, 18 (2009).
Acknowledgements
This project was supported by the National Natural Science Foundation of China (81773775, H.J., 82173857, H.J., 82373913 H.J.) and Shanghai Committee of Science and Technology (22S11900900, H.J.), Shanghai Frontiers Science Center of Optogenetic Techniques for Cell Metabolism and State Key Laboratory of Drug Research, SKLDR-2023-KF-04. We thank Prof. Kangdong Liu (Zhengzhou University) for technical assistance of PDX study. We thank Prof. Naixia Zhang (Shanghai Institute of Material Medica) for technical assistance of NMR study. We thank Prof. Hua Lu (Tulane University) for critical reading and editing of our manuscript. We thank Dr. Wenming Qin and Dr. Hai Wu at SSRF beamline BL19U for assistance with data collection. We thank the staff members of the Large-scale Protein Preparation System at the National Facility for Protein Science in Shanghai (NFPS), Zhangjiang Lab, Shanghai Advanced Research Institute, Chinese Academy of Science, China for providing technical support and assistance in data collection and analysis.
Author information
Authors and Affiliations
Contributions
This work was conceptualized by W.L. and J.H. PDX models and lung orthotopic models were performed by J.C., W.W., Q.H., T.G., Q.W., Y.Xue., X.Y. and H.J. Cell culture, qPCR, confocal immunoblot analysis, ChIP assay were done by J.C., Q.H., T.G., and Y.L. Immunoblot analysis, luciferase reporter, pull down and Co-IP experiments were performed by J.C., Q.H., X.L., and T.G. Plasmid construction, protein expression and purification were performed by T.G., and Y.L. X-ray co-crystal structure, NMR assay was performed by W.W., H.M., N.Z., Y. Xu., and W.L. Bioinformatics analysis were done by J.C., and Q.H. Data analysis and interpretation were done by W.L., J.C., W.W., Q.H., T.G., Y.L., H.M., B.F., Q.W., M.L., R.N., F.C., and J.H. The manuscript was drafted by W.L., J.C., W.W., Q.H., and J.H. and reviewed and modified by all the authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lu, W., Cui, J., Wang, W. et al. PPIA dictates NRF2 stability to promote lung cancer progression. Nat Commun 15, 4703 (2024). https://doi.org/10.1038/s41467-024-48364-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-48364-4