Abstract
Uncontrolled inflammatory response arising from the tumor microenvironment (TME) significantly contributes to cancer progression, prompting an investigation and careful evaluation of counter-regulatory mechanisms. We identified a trimeric complex at the mitochondria-associated membranes (MAMs), in which the purinergic P2X7 receptor - NLRP3 inflammasome liaison is fine-tuned by the tumor suppressor PML. PML downregulation drives an exacerbated immune response due to a loss of P2X7R-NLRP3 restraint that boosts tumor growth. PML mislocalization from MAMs elicits an uncontrolled NLRP3 activation, and consequent cytokines blast fueling cancer and worsening the tumor prognosis in different human cancers. New mechanistic insights are provided for the PML-P2X7R-NLRP3 axis to govern the TME in human carcinogenesis, fostering new targeted therapeutic approaches.
Similar content being viewed by others
Introduction
Promyelocytic Leukemia Protein (PML), a nuclear moonlighting protein with an established role in genetic control, also acts at the endoplasmic reticulum (ER)-mitochondria interface, namely, at mitochondria-associated membranes (MAMs) [1]. PML behaves as a tumor suppressor, regulating cell fate via Ca2+ transfer to mitochondria and thereby regulating cellular metabolism and death [2, 3].
Recently, MAMs were identified as platforms for inflammatory signaling that are regulated by the multiprotein complex NOD-like receptor family, pyrin domain containing-3 protein (NLRP3) inflammasome [4]. At rest, NLRP3 resides at the ER, whereas in response to danger signals it redistributes to MAMs together with its adapter ASC (apoptosis-associated speck-like protein), driving inflammasome assembly and maturation thus leading to IL-1β and IL-18 secretion [4, 5].
Inflammasome activation leads to defense against several pathogens, but lack of shutdown may be attributed to many diseases, including neurodegenerative, metabolic, and autoimmune diseases as well as cancer [6]. The negative feedback dampening NLRP3 inflammasome activation is poorly understood but essential [7].
The purinergic P2X7 receptor (P2X7R) is one of the most potent activators of the NLRP3 inflammasome. Well known to be a ubiquitous damage-associated molecular pattern (DAMP) receptor and to trigger the release of substantial amounts of IL-1β and IL-18 in response to extracellular ATP [8, 9], P2X7R was recently shown to interact with NLRP3 directly [10].
Here, we put forward the hypothesis of a new PML function in macrophages as a modulator of the P2X7R-NLRP3 axis at MAMs for a new understanding of the rational design of novel anti-inflammatory therapies.
Results
The PML/NLRP3/P2X7R complex at MAMs
Membranes at the ER-mitochondria interface (mitochondria-associated membranes; MAMs) have been reported as a platform for activating the NLRP3 inflammasome [4]. We previously reported that PML localizes at the ER and mitochondria contact sites, forming PML MAMs-bodies (PML-MaBs) and acting as a scaffold protein to regulate mitochondrial metabolism and cell death [2, 3]. Therefore, we hypothesized that PML at MAMs plays a role in controlling NLRP3 activation to trigger inflammation.
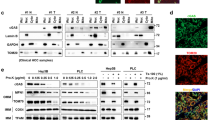
Co-immunoprecipitation (Co-IP) and proximity ligation assays (PLA) revealed close proximity of PML and NLRP3 in macrophages ER (Fig. 1A, B, and Supplementary Fig. S1A).
A Endoplasmic reticulum (ER) fraction obtained from the liver of wild-type (WT) mice was used for co-immunoprecipitation (Co-IP) of endogenous PML with NLRP3. Using PML as a bait, the levels of NLRP3 were detected, showing the interaction between NLRP3 and PML. B Proximity ligation assay (PLA) showing the interaction of NLRP3 with PML (red dots) in the ER fraction of WT resident peritoneal macrophages (pRMØs) left untreated or treated with LPS and nigericin. PDI (green) was used as an ER marker (scale bars, 20 μm). In the bottom panel, the zoomed regions display interaction sites at the ER (scale bar, 2 μm). In the upper right panel, the columns show the quantification of the PLA signal between NLRP3 and PML. Bars: mean ± SEM. (n = 3 independent experiments) C Confocal immunofluorescence staining images of NLRP3 (red) and calnexin (used as an ER marker, green) in WT and Pml−/− pRMØs untreated or treated with LPS and nigericin (scale bars, 5 μm). Merged images of the two immunostainings are shown. The right panel shows the proportion of clustered NLRP3 on the total NLRP3 signal. Bars: median. *p < 0.05. D Left panels show immunoblots of subcellular fractions isolated from WT and Pml−/− BMDMs. Where indicated, cells were exposed to LPS and nigericin treatment. The following markers were used: IP3R3 for the ER, VDAC for mitochondria (Mp, mito pure), Sigma 1-R for MAMs, and GAPDH for Cyt (cytosol). H: homogenate. Right panels show the quantification of the P2X7R and NLRP3 proteins in the ER and MAM fractions normalized to the amounts of IP3R3 and Sigma 1-R. The middle lines of each bar represent the mean values (n = 3 independent experiments). E ER fractions obtained from the livers of WT and Pml−/− mice were used for the Co-IP of endogenous NLRP3 with P2X7R. Using NLRP3 as bait, the presence of P2X7R was detected, showing that the interaction between NLRP3 and P2X7R depends on whether PML is lacking or after inflammasome activation. The lower panel shows the fold change of the NLRP3-P2X7R interaction; the middle lines of each bar represent the mean values (n = 3 independent experiments). F PLA shows the interaction between P2X7R and NLRP3 (red dots) in the ER of WT and Pml−/− pRMØs, untreated or treated with LPS and nigericin. PDI (green) was used as an ER marker. Scale bars, 20 μm. Higher magnifications show interaction sites at the ER (scale bars, 2 μm). In the right bar graphs, quantification of the number of PLA red dots per cell revealed an increased P2X7R-NLRP3 interaction in Pml−/− pRMØs. Data are shown as the mean ± SEM. Two independent experiments were carried out in triplicate. *p < 0.05, ****p < 0.0001. G Co-IP of endogenous PML and P2X7R in ER fraction obtained from WT mouse liver (as in a). Using PML as bait, the expression of P2X7R was detected, showing the interaction between P2X7R and PML. H Representative PLA (red dots) images of WT pRMØs untreated or treated with LPS and nigericin. PDI (green) was used as an ER marker. Scale bars, 20 μm. Higher magnifications show interaction sites at the ER (scale bars, 2 μm). The column graph on the right shows a quantitative analysis of the PLA signal between PML and P2X7R. Bars: mean ± SEM. Two independent experiments were carried out in triplicate.
We then investigated the ER localization of NLRP3 in wild-type (WT) and PML-depleted (Pml−/−) cells under both the resting condition and after treatment with the inflammasome inducers lipopolysaccharide (LPS) and nigericin. Co-immunostaining of NLRP3 and calnexin, an ER marker, revealed enhanced accumulation of NLRP3 at the ER in stimulated resident peritoneal macrophages (pRMØs) from Pml−/− mice (Fig. 1C).
The purinoceptor P2X7R is highly expressed within cells of the innate immune system, including macrophages, and its activation is a powerful signal to activate the NLRP3 inflammasome [11]. We thus examined whether P2X7R, in addition to its canonical plasma membrane expression, also localizes to the ER/MAM compartments, where NLRP3 resides and accumulates upon its activation, to mediate NLRP3 complex assembly and maturation.
We performed subcellular fractionation analyses of WT and Pml−/− bone marrow-derived macrophages (BMDMs) (Fig. 1D) at rest and after inflammasome stimulation with LPS and nigericin, focusing on the mitochondria, ER, and MAM fractions.
As expected, NLRP3 was detected in both the ER and MAM compartments, redistributing more to MAMs in Pml−/− cells than in WT cells after inflammasome activation (Fig. 1D). Consistently, we revealed a previously unknown localization of P2X7R at the ER/MAMs, wherein it accumulated considerably more in the absence of PML than in WT conditions (Fig. 1D). Furthermore, Co-IP and PLA revealed a stronger interaction between NLRP3 and P2X7R at the ER/MAMs in the absence of PML (Fig. 1E, F and Supplementary Fig. S1B).
Given the predominant role of PML as a scaffold protein, we examined its potential to interact with and function in complex with NLRP3 and P2X7R at the ER/MAMs.
PML, NLRP3, and P2X7R co-immunoprecipitated at the ER (Fig. 1A, E, G and Supplementary Fig. S1C), suggesting that they may form a trimer. Such findings were further confirmed by PLA, which showed a direct interaction between endogenous PML and endogenous P2X7R in ER macrophages (Fig. 1H), similar to what was observed for PML and NLRP3 (Fig. 1B).
The capability of NLRP3, P2X7R, and PML to form a trimolecular binding complex was assessed by qualitative surface plasmon resonance (SPR) measurements. In sequential injection experiments, we assessed the simultaneous binding of PML and P2X7R as analytes of NLRP3, immobilized on an optical sensor chip surface by GST tag capturing. The resulting sensorgrams (Supplementary Fig. S1D) showed that binding signals on the NLRP3 ligand were generated for both analytes, indicating a trimolecular interaction. The experiment shown in Supplementary Fig. S1E revealed that the two analytes PML and P2X7R were also able to interact with each other in the absence of NLRP3 (Supplementary Fig. S1E). Altogether, these data suggest that PML, NLRP3, and P2X7R work as triumvirates at MAMs, depicting a multiprotein complex in which PML acts as a negative regulator of NLRP3 function. In PML absence, NLRP3 and P2X7R lose their restraints and tighten together (Supplementary Fig. S1F).
PML tones down macrophage NLRP3 arousal
NLRP3 inflammasome activation and IL-1β release are thereby primarily triggered by macrophages, which are crucial for generating the inflammatory milieu that can sustain tumor growth [12]. We investigated whether the interaction among PML, P2X7R, and NLRP3 has functional consequences for NLRP3 activation and IL-1β production. pRMØs and BMDMs isolated from WT and Pml−/− mice and PML-silenced human peripheral blood mononuclear cells (PBMCs) (Supplementary Fig. S2A) were first primed with LPS and then stimulated with either the ATP analog benzoyl-ATP (Bz-ATP) or nigericin according to the established protocol to trigger NLRP3 activation and IL-1β release [13].
Lack of PML resulted in increased IL-1β release in response to both stimuli compared to that observed in the WT conditions (Fig. 2A–C). Consistently, the levels of caspase-1 p10 (an autoprocessed fragment of caspase-1) and cleaved IL-1β were higher in the media from activated Pml−/− pRMØs than in that from WT pRMØs (Fig. 2D and Supplementary Fig. S2B). Transduction of an ErPML chimera, which redistributed to only the MAM sites [2], into Pml−/− pRMØs ameliorated the IL-1β release to the levels measured in WT pRMØs, while the nuclear-targeted PML chimera was ineffective [2] (Supplementary Fig. S2C, D). Therefore, the localization of PML at MAMs indicates its relevance to repressing the activation of the NLRP3 inflammasome.
pRMØs (A) and BMDMs (B) from WT (red columns) and Pml−/− mice (black columns) were left untreated or stimulated with LPS and Bz-ATP or LPS and nigericin. The IL-1β levels in the supernatants were determined by ELISA. Error bars indicate the SEM of 3 independent experiments carried out in triplicate. *p < 0.05, **p < 0.01 C ELISA of mature IL-1β in the supernatants of siRNA-transfected PBMCs upon stimulation with LPS together with Bz-ATP. PBMCs were transfected with scramble or PML siRNA. Error bars indicate the SEM of 3 independent experiments carried out in triplicate. **p < 0.01. D The culture media and total cell lysates of pRMØs from WT and Pml−/− mice were collected before and after activation with LPS plus ATP or LPS and nigericin and were resolved by SDS–PAGE. The results shown are representative of 3 independent experiments. ELISA of IL-1β from peritoneal exudate (E) and blood (F) samples from WT (red columns) and Pml−/− mice (black columns) that were untreated or treated with LPS plus nigericin. Bars: mean ± SEM of 3 independent experiments carried out in triplicate. **p < 0.01, ***p < 0.001. G ELISA of IL-1β in Pml−/− pRMØs stimulated with LPS and Bz-ATP after transfection with scramble, siRNA NLRP3 and siRNA NLRC4. Bars: mean ± SEM of 3 independent experiments. **p < 0.01. H WT (red) and Pml−/− (black) C57BL/6 mice (7 per group) were injected subcutaneously with B16-F10 melanoma cells (1 × 106). The left graph shows tumor growth for 2 weeks after the injection. The middle graphs show the quantified tumor volumes and weights at 14 days post-injection. Error bars indicate SEM. **p < 0.01, ****p < 0.0001. The right panel shows representative tumors excised at the same time point (scale bar: 1 cm). I Immunohistochemistry (IHC) revealed much higher IL-1β immunoreactivity (IR) in the peritumoral areas of melanoma-bearing Pml−/− mice than in that of WT mice. TM: tumor mass. Nuclei were counterstained with Harris’ hematoxylin (HH). Scale bars: 50 μm. J Pml−/− C57BL/6 mice were injected subcutaneously with B16-F10 melanoma cells (1 × 106). An experimental group (green; n = 6) was injected IP with IL-1β antiserum (50 µg twice weekly; clone B122; BioXCell), while a blank group (black; n = 6) was injected IP with equal amounts of isotope control antibodies. The left graph shows tumor growth for 14 days after the injection. The middle graphs show the quantified tumor volumes and weights at 14 days post-injection. Error bars indicate SEM. **p < 0.05, ****p < 0.001. The right panel shows representative tumors excised at the same time point (scale bar: 1 cm). K IHC revealed an almost complete lack of IL-1β-IR in the peritumoral areas of melanoma-bearing Pml−/− mice treated in vivo with IL-1β antiserum compared with those of the blank group. TM tumor mass. Nuclei were counterstained with HH. Scale bars: 50 μm.
To validate these in vitro results, we directly treated WT and Pml−/− mice with LPS in combination with nigericin by intraperitoneal (IP) injection and measured the level of IL-1β 4 h later. The release of IL-1β into the peritoneal exudate, blood, and liver as well as the caspase-1 activity, were strongly increased in the Pml−/− mice compared with the WT mice (Fig. 2E, F and Supplementary Fig. S2E, F). Pharmacological inhibition of NLRP3 activation by MCC950 (a known NLRP3 inhibitor) [14] or AZ10606120 dihydrochloride (a known P2X7R inhibitor) [15] counteracted the IL-1β increase in Pml−/− mice both in vitro and in vivo (Supplementary Fig. S2G–J).
To further determine whether PML is capable of controlling the activation of NLRC4 (CARD12, IPAF), another inflammasome complex known to drive IL-1β release [16], we genetically interfered with NLRC4 and NLRP3 in Pml−/− pRMØs. Unlike silencing NLRP3, NLRC4 silencing did not attenuate IL-1β secretion in response to stimulation, indicating that PML is involved in specifically suppressing NLRP3-dependent inflammation (Fig. 2G and Supplementary Fig. S2K). Accordingly, the NLRP3-stimulated secretion of IL-18 was also upregulated in the absence of PML (Supplementary Fig. S2L), whereas this effect was not observed for other pro-inflammatory cytokines independent of NLRP3 stimulation, such as IL-6 (Supplementary Fig. S2M) and TNFα (data not shown).
Altogether, these data suggest that PML is a major driver of NLRP3 and P2X7R localization at MAMs, limiting their capacity to activate the inflammasome.
PML inactivation boosts tumor growth
It is well known that sustained activation of the NLRP3 inflammasome promotes tumorigenesis at several stages [6, 17]. Hence, high levels of IL-1β have been implicated in carcinogenesis and tumor progression [18].
We investigated whether a Pml−/− host, which was skewed toward NLRP3-dependent inflammation, would provide a more permissive environment for tumor proliferation than a WT host. We compared the growth of B16-F10 melanoma and LL/2 Lewis lung carcinoma syngeneic cell lines, which express endogenous PML (Supplementary Fig. S3A). These cells were injected subcutaneously into either WT or Pml−/− mice. Tumor growth was greatly accelerated in Pml−/− mice (Fig. 2H and Supplementary Fig. S3B, C) and associated with the hypersecretion of IL-1β in peritumoral tissues (Fig. 2I and Supplementary Fig. S3D) compared with that observed in WT mice.
Cancer cell proliferation is stimulated by inflammation [19]. To assess whether the accelerated tumor progression in the Pml−/− host was dependent on enhanced IL-1β secretion and thus on increased NLRP3 activation, we used an anti-IL-1β serum to deplete the cytokine in the TME. IL-1β immunoblockade dampened tumor growth in the Pml−/− hosts (Fig. 2J, K) without significantly affecting tumor development in WT hosts (Supplementary Fig. S3E). Thus, a fertile environment for accelerated tumor growth in PML-null hosts is generated by the increased release of IL-1β into the TME. Accordingly, knocking down the IL-1β receptor (IL-1R1) in B16-F10 melanoma cells by CRISPR/CAS9 (Supplementary Fig. S3F, G) reduced both B16-F10 melanoma cell proliferation and the tumor size in the Pml−/− hosts (Supplementary Fig. S3H), highlighting the fundamental tumor-promoting role of IL-1β.
PML deficiency drives M2 polarization in the TME
Macrophages that infiltrate tumor tissues, also known as tumor-associated macrophages (TAMs), are the major component of the highly complex TME, in which cancer cells coexist with immune and nonimmune cells [20]. TAMs preferentially express an M2-like phenotype and promote tumor growth [21].
We investigated whether PML-dependent activation of the NLRP3 inflammasome contributes to the polarization of macrophage phenotypes within the TME.
Flow cytometry revealed no significant differences in the percentages of both Ly6C+ and Ly6C- monocytes between WT and Pml−/− mice under resting conditions (Supplementary Fig. S4A), indicating no variation in the number of these circulating cells. We then investigated whether the different amount of IL-1β released into the TME by WT and Pml−/− mice was dependent on the M1/M2 polarization of TAMs. Immunohistochemical and FACS analyses (Supplementary Fig. S4B for gating strategy) revealed in Pml−/− mice a significant shift in TAMs toward the M2 phenotype compared with WT mice, with the loss of M1-associated expression markers (Fig. 3A–C).
A Representative tumor sections excised from WT (upper panels) and Pml−/− mice (lower panels), stained with H/E or immunostained for F4/80, iNOS, CD206 (mannose receptor; manR) and Ly6g; nuclei were counterstained with HH. Note the high number of free cells in the peritumoral areas and their larger occurrence in Pml−/− mice. TM: tumor mass. Scale bars: 50 µm. B The peritumoral tissue of Pml−/− mice (black columns) is distinctly enriched with F4/80-, mannose receptor- and Ly6g-immunoreactive cells and almost devoid of iNOS-immunoreactive cells compared with that of WT mice, depicting an immunosuppressive environment. The data are expressed as the mean ± SEM. Significantly different from WT mice *p < 0.05, **p < 0.01, ****p < 0.0001. C Absolute number of TAMs (CD3- CD45R- F4/80+ CD11b+)/tumor mass from WT and Pml−/− mice (n = 7 per group). TAM polarization was analyzed by measuring CD86 and MHC-II expression based on the mean fluorescence intensity (MFI). Left panels: the dots indicate the results of individual mice, and the lines indicate the mean values. **p < 0.01. Right panels: representative histogram plots from 2 mice/group of CD86 and MHC-II expression in macrophages. D Analysis of tumor growth after bone marrow replacement. WT mice were lethally irradiated to induce myeloablation and then transplanted with bone marrow from WT (PML-WT > PML-WT) or Pml−/− (PML-KO > PML-WT) animals. Bone marrow reconstitution was monitored for 16 weeks (Supplementary Fig. S4 and Methods section), then chimeric mice were subcutaneously injected with 1 × 106 B16-F10 melanoma cells. Tumor growth was monitored at the indicated time points. Numerical results are expressed as the mean ± SEM (n = 4 per group). E Co-culture with activated Pml−/− macrophages showed a larger tumor cell growth-promoting effect compared to that exerted by WT macrophages. B16-F10 cells were co-cultured with pRMØs from WT and Pml−/− mice and treated daily with LPS and Bz-ATP. After one week, the proliferation of B16-F10 cells was analyzed by crystal violet staining (595 nm absorbance). Experiments were performed twice, and the error bars indicate SEM. ****p < 0.0001.
In most cancer types, TAMs are recruited from bone marrow progenitors [22, 23]. To verify the essential non-cell-autonomous role of Pml−/− immune cells in providing a tumor-supportive microenvironment, we generated bone marrow chimeras (Supplementary Fig. S4C–F). We injected subcutaneously B16-F10 melanoma syngeneic cells into WT mice transplanted with either WT or Pml−/− bone marrow-derived mononuclear cells (BMMNCs) (Fig. 3D). Tumor growth was significantly faster and greater in mice transplanted with Pml−/− cells than in those transplanted with WT BMMNCs (Fig. 3D), highlighting the role of PML-deficient hematopoietic cells in supporting increased tumor growth.
Next, to assess the direct involvement of TME macrophages in supporting cancer cell proliferation along the PML/P2X7R/NLRP3 axis, we established a co-culture model. pRMØs from WT and Pml−/− mice were cultured in removable inserts on the top of the plate, activated by LPS and Bz-ATP as inflammasome activators, and incubated with B16-F10 melanoma cancer cells seeded on the bottom of the plate; the cells were separated by a porous membrane that allowed the free exchange of soluble factors and cytokines (Supplementary Fig. S5A). Co-culture with activated Pml−/− macrophages exerted a larger tumor cell growth-promoting effect compared to that of WT macrophages (Fig. 3E). Direct stimulation of cancer cells with LPS had no effect on their growth (Supplementary Fig. S5B). Pretreatment with anti-IL-1β serum before macrophage activation or IL-1R1 knockdown considerably reduced the growth of cancer cells (Supplementary Fig. S5C, D). Accordingly, the H460 human lung cancer cell line in contact with activated PML-silenced PBMCs showed an increased growth rate compared with that of control PBMCs (Supplementary Fig. S5E). Altogether, these results suggest that PML loss in macrophages favors tumor establishment and progression by enhancing IL-1β secretion from M2-polarized TAMs.
The PML/NLRP3 axis in human cancers
After elucidating the PML-dependent NLRP3 activation pathway, we next explored its clinical relevance in human cancers. Two retrospective cohorts of patients diagnosed with cutaneous melanoma or non-small-cell lung cancer (NSCLC) (see Table S1) were selected. Within these cohorts, we distinguished two groups of patients based on 5-year follow-up data: patients with long survival and NO survival times. Immunohistochemical analysis on melanoma and NSCLC tissue samples (Fig. 4A and Supplementary Fig. S6A) revealed that, compared with the long-survival group, the short-survival patients had more CD68- and CD206-immunoreactive TAMs and greater IL-1β immunoreactivity in the peritumoral stroma (Fig. 4B, C and Supplementary Fig. S6B, C), and these parameters were highly correlated. However, the number of PML-immunoreactive cells did not differ significantly between the two observation groups (data not shown). Indeed, other parameters were carefully evaluated and proven to be very informative, such as (i) the positive linear correlation between inflammatory signs (e.g. number of free cells recruited to the TME) and the IL-1β level in the peritumoral area observed in individual tumor specimens and (ii) the inverse correlation between nuclear and extranuclear PML localization and the release of IL-1β into the TME (Fig. 4A–D and Supplementary Fig. S6A–D).
A Histological sections of human melanoma tissue samples representative also of the peritumoral tissue and belonging to long-survival (up to 5 years) and short-survival (less than 5 years) patients were immunostained for CD68, CD206, IL-1β and PML (see labels). Representative areas are shown at higher magnification to detail the protein expression in macrophages (CD206; arrows) and the differential subcellular localization of PML (arrows). IL-1β levels were closely correlated with the occurrence of CD68+ TAMs, the differential subcellular localization of PML, and poor prognosis. B Patient survival was negatively correlated with TAM M2 polarization and IL-1β expression. C Il-1β expression levels in human melanoma tissue samples and 5-year survival rates. D The nuclear localization of PML was predictive of poor prognosis for melanoma patients.
The long-survival group achieved a complete response (CR) without any recurrences, whereas all patients in the short-survival group died within 5 years after diagnosis (Fig. 4C and Supplementary Fig. S6C). IL-1β levels were closely correlated with the numbers of CD68+ or CD206+ TAMs (Fig. 4B and Supplementary Fig. S6B) and might indeed be predictive of poor prognosis after the onset of melanoma or NSCLC (Fig. 4C and Supplementary Fig. S6C). The most important discovery was that PML exhibited differential subcellular localization inside the TAMs in long- and short-survival patients’ tissue samples, with mostly cytoplasmic localization being observed in the former group and nuclear localization being observed in the latter (Fig. 4A, D and Supplementary Fig. S6A, D).
Altogether, the data from human samples likely confirm those obtained from mouse models and indicate that a shift in the subcellular localization of PML from an extranuclear region to the TAMs nuclei drives their M2 polarization and promotes tumor development boosted by excessive inflammation.
P2X7R/NLRP3 targeting counteracts PML-loss driven tumorigenesis
To determine whether targeting NLRP3 could prevent the generation of pro-inflammatory, and tumor-promoting stimuli derived from Pml−/− macrophages, we used three different strategies in Pml−/− mice: (i) NLRP3 blockade by the selective inhibitor MCC950; (ii) genetic deletion of NLRP3; and (iii) genetic deletion of P2X7R, the most powerful NLRP3 activator (Supplementary Fig. S7A, B). Consistent with a reduction in the PML/NLRP3/IL-1β axis, we observed lower amounts of IL-1β in pRMØ, BMDM, peritoneal exudate, and blood samples collected from Pml−/−/P2x7r−/− and Pml−/−/Nlrp3−/− double-knockout (DKO) mice than in those collected from Pml−/− mice (Fig. 5A–D).
pRMØs (A) and BMDMs (B) from Pml−/−, Pml−/−/P2x7r−/− and Pml−/−/Nlrp3−/− mice were left unstimulated or stimulated with LPS plus Bz-ATP. The IL-1β levels in the supernatants were determined by ELISA. Bars: mean ± SEM of 3 independent experiments carried out in triplicate. ****p < 0.0001. ELISA of IL-1β levels in peritoneal exudate (C) and blood (D) samples from Pml−/−, Pml−/−/P2x7r−/− and Pml−/−/Nlrp3−/− mice after treatment with LPS and nigericin. Bars: mean ± SEM of 3 independent experiments carried out in triplicate. ***p < 0.001. ****p < 0.0001. E Pml−/− (black line) and Pml−/−/Nlrp3−/− (purple line) C57BL/6 mice (6 per group) were injected subcutaneously with B16-F10 melanoma cells (1 × 106). Left panel, the graph shows the tumor kinetics at the indicated time points. The middle panels show the quantified tumor volumes and tumor weights at 14 days post-injection. Error bars indicate SEM. **p < 0.01, ****p < 0.0001. The right panel shows representative tumors excised at the same time point (scale bars: 1 cm). F. Pml−/− (black line) and Pml−/−/P2x7r−/− (gray line) C57BL/6 mice (6 per group) were injected subcutaneously with B16-F10 melanoma cells (1 × 106). Tumor growth was analyzed as described in e. Error bars indicate SEM. *p < 0.05, **p < 0.01, ***p < 0.001. Scale bar: 1 cm. G Representative tumor sections from specimens of different mouse groups: Pml−/− (upper panels), Pml−/−/Nlrp3−/− (middle panels) and Pml−/−/P2x7r−/− (lower panels). The sections were stained with H/E or immunostained for IL-1β, F4/80, iNOS, mannose receptor (manR) and Ly6g. Nuclei were counterstained with HH. TM: tumor mass. Bars: 50 µm. It is worth noting the massive recruitment of free cells and IL-1β immunoreactivity in the peritumoral areas of Pml−/− mice, which was otherwise hardly detectable in double-knockout mice. H Quantification of the cell density (N/mm2) in peritumoral tissues revealed significantly higher numbers of F4/80-, mannose receptor-, and Ly6g-immunoreactive cells in Pml−/− mice than in Pml−/−/P2x7r−/−and Pml−/−/Nlrp3−/− mice, while no significant differences were found in the numbers of iNOS-immunoreactive cells among the groups. Error bars indicate SEM *p < 0.05, ***p < 0.001, ****p < 0.0001.
Moreover, genetic or pharmacological inhibition of P2X7R or NLRP3 drastically reduced B16-F10-induced melanoma growth driven by PML deficiency (Fig. 5E, F and Supplementary Fig. S7C).
Interestingly, a significant reduction in the levels of secreted IL-1β, a reduced number of tumor – infiltrating macrophages, and a decreased ratio of M2-like macrophages in peritumoral tissues were observed after NLRP3 inhibition compared with those in Pml−/− mice (Fig. 5G, H). These results strongly indicate a crucial role of NLRP3 in M2 polarization driven by PML loss, which leads to enhanced tumorigenesis.
Discussion
PML is a tumor suppressor that helps to control gene expression by associating with nuclear bodies (NBs). While PML exerts its main effects in the nucleus, we showed that it is also expressed at MAMs, the region in which the ER and mitochondria are juxtaposed [2]. In this cellular compartment, PML determines the fate of the cell, controlling cell metabolism or death, by modulating Ca2+ transfer to mitochondria [2, 3]. In particular, we proposed that PML forms NBs-like structures (PML-MaBs) at the ER-mitochondria contact sites, where it orchestrates various functions by interacting with several proteins and generating multiprotein complexes. Interestingly, MAMs have been shown to function as a platform for inflammatory signaling regulated by the most widely characterized inflammasome, NLRP3 [4]. Two previous studies revealed a possible liaison between PML and NLRP3 [24, 25]. The earlier study demonstrated that PML could promote NLRP3 activation [24], while the latter proposed PML to be a novel regulator of ASC nuclear localization, thereby limiting NLRP3 activation [25]. To date, the mechanisms that control the formation and activation of NLRP3 at MAMs are unknown, as is the role of PML in this cell compartment. It is worth elucidating this PML-mediated inflammatory response due to obvious clinical implications.
In this study, we provide evidence that PML at MAMs represses NLRP3 activation in the context of a molecular triumvirate that includes the purinergic P2X7R. In the ER/MAM compartments, PML has been reported to directly interact with both NRLP3 and P2X7R. These three proteins form a multiprotein complex in which PML plays a fundamental role in counteracting NLRP3 activation mediated by P2X7R. The loss of PML promotes a NLRP3-related cytokine storm in response to stress conditions due to the increased redistribution and interaction of NLRP3 and P2X7R at MAMs. This mechanism is consistent with the hypothesis that PML is involved in retaining ASC, the adapter protein required for proper NLRP3 assembly, and the prevention of inflammasome hyperactivation, in the nucleus. In fact, PML deficiency may lead to higher redistribution of ASC in the cytoplasm, wherein it initiates inflammasome assembly at ER-related compartments together with NLRP3 [25].
We thus revealed that PML loss might boost tumorigenesis via the NLRP3-mediated fueling of the inflammatory response. Indeed, PML deficiency in the TME favors tumor nesting and development by increasing IL-1β secretion. This in turn promotes tumor growth, exerting a direct effect on cancer cells. Tumor growth was strongly reduced by inhibiting host NLRP3, acting on P2X7R, a major inflammasome activator, or directly impeding NLRP3 assembly.
Macrophages are the main immune cell type that specifically populates the TME and are highly prevalent in inflammation-sustained tumors. We found that PML-deficient macrophages under stress conditions were skewed toward the M2 phenotype and exacerbated NLRP3-mediated inflammation, exerting a promotional effect on tumor growth via the hypersecretion of IL-1β. Recent findings have indeed revealed the powerful pro-inflammatory potential of M2 macrophages in pancreatic and liver cancer [26, 27], although earlier data primarily associated them with anti-inflammatory action (in contrast to M1). Consistent with our observation from syngeneic mouse models, we found an inverse correlation between PML and IL-1β amounts in the peritumoral areas of patient specimens, and a higher rating of PML inside the nucleus in those with a bad prognosis. Further studies are needed to elucidate the mechanisms that modulate PML localization and expression in the TME. The recent identification of a new PML ubiquitination and degradation pathway [28] supports our findings that host PML loss may drive an immune response that worsens rather than ameliorates cancer, thereby enhancing IL-1β release and opening a new route for PML protein stabilization.
In summary, we identified a novel function of PML in undermining the P2X7R/NLRP3 liaison in ER/MAMs. This notion highlights the importance of these structures, first described by our research group and named PML-MaBs. We demonstrated how PML-deficient macrophages would drive tumor progression (in association with poor prognosis) by hyperactivating the IL-1β-driven inflammatory response. Our study thus elucidated pleiotropic tumor-promoting mechanisms due to PML loss, and expanded to outside the tumor, in the TME (Supplementary Fig. S7D). In addition, we suggest that the nuclear localization of PML should be evaluated (with simple, rather inexpensive, and routine analyses) in larger cohorts to determine whether it is predictive of poor prognosis in patients with melanomas, NSCLC, and possibly other solid tumors.
Finally, our findings may have implications for severe inflammatory syndromes including pathogen-related ones (as those related to COVID-19) and tissue specific ones (as neuroinflammation) for the following reasons. (i) Several viruses have been shown to disrupt PML-NBs [29]; the same could also happen to PML-MaBs, with consequent mislocalization of PML from the ER/MAM region, which would lead to enhanced IL-1β release; (ii) the induction of IL-1β by herpes simplex virus (HSV-1) was markedly enhanced in PML-deficient BMDMs [25]; this fueling of IL-1β release could presumably also be induced by other pathogens, as SARS-Cov-2, in PML-deficient macrophages; (iii) in brain pathologies NLRP3 dysregulation promotes the neuroinflammatory process exacerbating neuronal death [30]; in this context, NLRP3 driven inflammation might be supported by PML-deficient microglia.
The following key questions remain: (i) Can PML-MaBs disorders determine inflammation-related diseases severity (COVID-19 and neurodegenerative disorders)? (ii) Can PML localization at MAMs play a fundamental role in limiting inflammasome activation in response to pathogens (such as SARS-Cov-2)? We believe that these questions are worthy of experimentation. The models we propose might indeed pave the way for new studies on the mechanisms underlying severe inflammatory diseases.
Methods
Cell culture and transfection
B16-F10 melanoma cells (kindly provided by Pier Paolo Di Fiore’s lab at IFOM) were grown in RPMI medium supplemented with nonessential amino acids (Sigma-Aldrich), 10% fetal bovine serum (FBS), and 1% penicillin/streptomycin (P/S). The LL/2 mouse Lewis lung carcinoma cell line (from Sigma) and H460 cells (kindly provided by the Pagano laboratory [31]) were cultured in Dulbecco’s modified Eagle’s medium containing 10% FBS, 1% glutamine, and 1% P/S. Bone marrow-derived macrophages (BMDMs) from WT, Pml−/−, Pml−/−/P2rx7−/− and Pml−/−/Nlrp3−/− mice were isolated as described [32] and differentiated for 7 days in Iscove′s modified Dulbecco′s medium (IMDM) supplemented with 15% FBS (Gibco), 1% glutamine, 1% P/S and 40 ng/ml M-CSF. Resident peritoneal macrophages (pRMØs) from WT, Pml−/−, Pml−/−/P2rx7−/− and Pml−/−/Nlrp3−/− mice were isolated as described [33] and cultured in IMDM supplemented with 15% FBS (Gibco), 1% glutamine and 1% P/S. pRMØs from Pml−/− mice were transfected with siRNAs targeting NLRP3 and NLRC4 using Lipofectamine RNAiMAX (Invitrogen) or with the erPML or nuPML chimeric plasmid using jetPEI-Macrophage (Polyplus transfection). Human PBMCs were isolated as described [34], cultured in RPMI medium supplemented with nonessential amino acids (Sigma-Aldrich), 10% FBS and 1% P/S, and transfected with a siRNA targeting PML using Lipofectamine RNAiMAX (Invitrogen). All cells were grown in a 5% CO2 incubator at 37 °C and tested with a Mycoplasma PCR Detection Kit from Sigma (MP0035-1KT).
Reagents
ELISA kits for mouse IL-1β, human IL-1β, and mouse IL-6 were purchased from R&D Systems, for mouse IL-18 was purchased from MBL. The anti-IL-1β (clone B122) was purchased from BioXCell. The PML, NLRP3 and NLRC4 siRNAs were purchased from Thermo Fisher Scientific. AZ10606120 dihydrochloride was purchased from Tocris; lipopolysaccharide (LPS) from Escherichia coli 055:B5 (L2880), 2′(3′)-O-(4-benzoylbenzoyl)adenosine 5′-trisphosphate triethylammonium salt 95% (Bz-ATP) (B6396), ATP (A3377) and nigericin sodium salt (N7143) were purchased from Sigma, and MCC950 (HY-12815A) was purchased from Invivogen. Caspase-1 assay kit was from Abcam.
Immunoblotting
Total cell lysates from BMDM, pRMØs, PBMCs, B16-F10, and LL/2 cells were lysed in a buffer containing 50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% Triton X-100, and 0.2% SDS supplemented with protease and phosphatase inhibitor cocktails. Protein extracts were quantified by the Lowry method, and 10 μg of proteins were loaded and separated on a NuPAGE Novex 4–12% Bis-Tris Gel (Life Technologies) and transferred onto a nitrocellulose membrane.
Primary antibodies were raised against human PML (Abcam, ab72137), mouse PML (Millipore, MAB3738), mouse PML (Abcam, #67761), the IL-1 receptor (I/IL-1R1, Abcam, ab107270), Sigma-1R (Sigma-Aldrich, HPA018002), IP3R3 (BD Biosciences, 610312), GAPDH (Cell Signaling, #2118), VDAC (Abcam, ab15895), β-tubulin (Cell Signaling, #55685), NLRP3 (Adipogen, AG-20B-0014-C100), P2X7 (Alomone, APR-004), P2X7 (Sigma-Aldrich, P8232), Caspase-1 (Novus Biological, #14F468), IL-1β (Cell Signaling Technology, #12242), and β-Actin (Sigma-Aldrich, #A2668). HRP-conjugated antibodies (#31430; #31460. WB, 1:5,000 from Thermo Fisher Scientific) were used, followed by detection using ImageQuant LAS 4000 (GE Healthcare) (see Supplemental Material). The western blots shown in the figures are representative of at least 3 independent experiments.
Inflammasome stimulation
BMDMs were seeded at a density of 5 × 105 cells per well in 24-well plates, while pRMØs were seeded at 3 × 105 and 1 × 106 cells per well in 24- and 6-well plates, respectively. Human PBMCs were seeded at a density of 5 × 105 cells per well in 24-well plates. The next day, the overnight (O/N) medium was replaced, and cells were stimulated with LPS (1 μg/mL) in fresh medium for 2 h and then stimulated with one of the following inflammasome activators: 5 mM ATP (1 h), 100 μM Bz-ATP (30 min) or 10 μM nigericin (1 h).
Supernatants were assayed by ELISA according to the manufacturer’s instructions (R&D Systems or MBL). The data shown in the figures are representative of at least 3 different independent experiments.
Analysis of cleaved Caspase-1 and cleaved IL-1β levels in conditioned cell culture media
A total of 1 × 106 pRMØs from WT and Pml−/− mice were cultured in complete IMDM. The next day, the O/N medium was replaced, and the cells were stimulated with LPS (1 μg/mL) for 3 h and then with the inflammasome activator 5 mM ATP (1 h) or 10 μM nigericin (1 h). The conditioned cell culture media were then collected and concentrated using Pierce Protein Concentrators PES 10 K MWCO (Thermo Fisher) and centrifuged (3000 g for 15 min). Aliquots of media (10 μl) were analyzed by SDS–PAGE and immunoblotting (Ponceau S staining was used as a loading control). The levels of cleaved Caspase-1 and cleaved IL-1β were detected with anti-Caspase-1 (Novus Biological, #14F468) and anti-IL-1β (Cell Signaling Technology, #12242) antibodies, respectively.
Gene knockout by CRISPR/Cas9
The procedure was performed in accordance with a previously published protocol [35]. To prevent off-target effects, RNA guides (gRNAs) were purchased from Integrated DNA Technologies IDT. The 20-nucleotide RNA guide sequence was transfected into B16-F10 cells with Streptococcus pyogenes Cas9 (IDT) using Lipofectamine RNAiMAX Transfection Reagent (Thermo Fisher). After two days of incubation, single cells were isolated by serial dilution and expanded for 5 weeks. Gene KO was assessed by WB and confirmed by RT–PCR.
Co-culture assay
pRMØs from WT and Pml−/− C57BL/6 mice were seeded in 6-well plates until 80% confluence and co-cultured in an insert chamber placed on top of the B16-F10 cells. IL-1R1 KO B16-F10 cells generated using the CRISPR/Cas9 technique were co-cultured with pRMØs from Pml−/− mice. The bottom of the insert chamber had 0.4-μm pores, allowing cytokines and growth factors produced by pRMØs to reach the lower chamber containing B16-F10 cells or IL-1R1 KO B16-F10 clones. The cells were cultured for up to 1 week, and pRMØs were treated daily with LPS (1 μg\ml) plus Bz-ATP (100 μM) in the absence or presence of anti-IL-1β (50 pg/ml). Equal amounts of PBS or the isotype control antibody were used as controls. H460 cells were co-cultured with human PBMCs transfected with the scramble or PML siRNA and treated daily with LPS and Bz-ATP. After one week, the inserts were removed, and the tumor cells (B16-F10, IL-1R1 KO B16-F10 clones and H460 cells) were stained with crystal violet. After lysis with 10% acetic acid, the absorbance was read at 595 nm.
Subcellular fractionation
WT and Pml−/− C57BL/6 mice were treated with LPS (250 μg/Kg IP for 1 h) and nigericin (2.5 mg/kg IP for 1 h), and their livers were surgically removed after anesthetic overdose. BMDM from WT, Pml−/− were obtained and isolated as described above and were treated with LPS (1 μg/mL) for 2 h and then stimulated with 10 μM nigericin (1 h).
Fractionation was performed as described previously [36, 37]. Briefly, BMDM (109) were harvested, washed by centrifugation at 500 × g for 5 min with PBS, and resuspended in homogenization buffer (225 mM mannitol, 75 mM sucrose, 30 mM Tris-HCl pH 7.4, 0.1 mM EGTA and PMSF) and gently disrupted by Dounce homogenization. Livers were fragmented and homogenized in homogenization buffer (225 mM mannitol, 75 mM sucrose, 30 mM Tris-HCl pH 7.4, 0.5 mM EGTA and 0.5% BSA).
The homogenate was centrifuged twice at 600 × g for 5 min, then the supernatant was centrifuged at 10,000 × g for 10 min to pellet crude mitochondria. The resultant supernatant was centrifuged at 20,000 × g for 30 min at 4 °C. Further centrifugation of the obtained supernatant at 100,000 × g for 90 min (70-Ti rotor, Beckman, Milan, Italy) at 4 °C results in the isolation of cytosolic fraction (supernatant) and ER (pellet). The crude mitochondrial fraction was resuspended in isolation buffer (250 mM mannitol, 5 mM Hepes (pH 7.4) and 0.5 mM EGTA), subjected to Percoll gradient centrifugation (Percoll medium: 225-mM mannitol, 25-mM HEPES (pH 7.4), 1-mM EGTA and 30% Percoll (vol/vol) at 95,000 × g for 30 min (SW40 rotor). A dense band containing purified mitochondria was recovered approximately at the bottom of the ultracentrifuge tube, washed by centrifugation at 6300 × g for 10 min, and resuspended in isolation buffer. The MAMs fraction was removed from the Percoll gradient, diluted in isolation buffer, and centrifuged at 100,000 × g for 90 min (70-Ti rotor); the pellet was the MAMs fraction. IP3R3, Sigma 1-R, GAPDH, and VDAC were used as markers of the ER, MAMs, cytosol, and pure mitochondria, respectively.
In vivo LPS challenge
WT, Pml−/−, Pml−/−/P2rx7−/− and Pml−/−/Nlrp3−/− C57BL/6 mice were treated with LPS (250 μg/kg IP for 4 h) and nigericin (2.5 mg/kg IP for 1 h), and their blood and peritoneal exudates were collected after anesthetic overdose. Pml−/− mice were pretreated with MCC950 (20 mg/kg, IP), AZ10606120 (5 mg/kg, IP) or vehicle (PBS, IP) 30 min before treatment with LPS (250 μg/Kg IP for 4 h) and nigericin (2.5 mg/kg IP for 1 h). Thereafter, their blood and peritoneal exudates were collected after anesthetic overdose. The peritoneal exudate samples were concentrated using Pierce Protein Concentrators PES 10 K MWCO (Thermo Fisher), and the supernatant was centrifuged (3000 × g for 15 min). The concentrated medium (10 μl) was separated by SDS/PAGE and transferred onto a nitrocellulose membrane for standard WB. For the caspase-1 assay, WT and Pml−/− C57BL/6 mice were treated with LPS (250 μg/kg IP for 1 h) and nigericin (2.5 mg/kg IP for 1 h). Their livers were removed after an anesthetic overdose and homogenized with a Potter pestle in lysis buffer (300 mM sucrose, 1 mM K2HPO4, 5.5 mM D-glucose, 20 mM HEPES, 1 mM phenylmethylsulfonylfluoride, and 0.5% IGEPAL). Tissue extracts were then centrifuged (12,000 × g at 4 °C for 15 min). Protein extracts (200 μg) were assayed for caspase-1. Mouse plasma was collected after blood centrifugation (1000 × g, 10 min at 4 °C), and the IL-1β levels were determined by ELISA according to the manufacturer’s instructions.
Tumor generation and in vivo drug administration
Procedures involving animals and their care were in conformity with institutional guidelines, and the Animal Ethics Committee approved all experimental protocols (authorizations n. 481/2017-PR and CBCC2.N.BH4 approved by the Italian Ministry of Health). To generate DKO C57BL/6 mice, the Pml−/− mice were crossed either with the P2rx7−/− strain (a gift from F. Di Virgilio) or the Nlrp3−/− strain. All mice were housed in a temperature-controlled environment on a 12 h light/dark cycle and received food and water ad libitum. A total of 1 × 106 B16-F10 melanoma cells, IL-1R1 KO B16-F10 clones or LL/2 cells were injected subcutaneously into different groups of 6- to 8-week-old C57BL/6 female mice. Tumor growth was monitored daily, and tumor volumes were measured with calipers according to the following equation: Volume = π/6 × (a × b2), where a is the major diameter and b is the minor diameter. The mice were randomly divided into the treatment and respective control groups (5–6 mice per group). For NLRP3 inflammasome inhibition, mice were treated with MCC950 (20 mg/kg IP) or AZ10606120 (5 mg/kg intratumor) three times a week. For the cytokine neutralization experiments, mice were treated with anti-IL-1β serum (BioXCell, clone B122) (50 µg IP, twice weekly). Controls were treated IP with an equal amount of isotype control antibodies or an equal volume of PBS. After an anesthetic overdose, tumors were surgically removed from all mice that reached the experimental endpoint, after which they were weighed and trimmed for immunohistochemistry or FACS analyses.
Histology and immunohistochemistry
Tumor masses, including peritumoral stroma, were collected at 14 days postinoculation, fixed in Bouin’s liquid for 7 h at 4 °C, dehydrated in a cold-graded ethanol series and embedded in paraffin. For each specimen, at least ten sets (50 µm away) of five serial sections (6 µm thick) were stained with Harris’ hematoxylin and eosin (H/E) for general histology or processed for immunohistochemistry, which was performed using ABC-peroxidase. Adjacent sections were incubated for 1 h at room temperature with antibodies against F4/80 (CI: A3-1) (Santa Cruz Biotech; sc-59171), IL-1β (H-155) (Santa Cruz, sc-7884), iNOS (Abcam, ab15323), mannose receptor antibody (Abcam, ab64693) and Ly6g [RB6-8C5] (Abcam, ab25377). The antibodies were diluted 1:10 to 1:50 in 0.01 M PBS (pH 7.3) containing 0.1% sodium azide. A hybridoma irrelevant supernatant or normal rabbit serum was substituted for the primary antibody in the negative controls. The antibodies were tested on sections after or without antigen retrieval steps (40 min at 95 °C in 0.01 M, pH 6.0 sodium citrate buffer containing 0.05% Tween 20, followed by cooling at RT and rinsing with PBS Tween 20). Thereafter, the sections were incubated for 60 min with biotinylated goat anti-rat IgG (mouse adsorbed) or biotinylated goat anti-rabbit IgG sera (Vector Labs., Burlingame, USA) and diluted 1:200 or 1:1000, respectively, with PBS containing 0.1% sodium azide and 1% BSA. Some sections (not treated with any primary antibodies) were incubated with biotinylated horse anti-mouse IgG serum (Vector) diluted 1:1000 to label B cells. Thereafter, the sections were incubated for 60 min with avidin-biotinylated peroxidase complex (ABC, Vectastain Elite, Vector). Following rinsing and staining (diaminobenzidine), the sections were slightly counterstained with Harris’ hematoxylin, rinsed, dehydrated, mounted and examined under bright-field illumination.
Immunoreactive cells (nucleated only) in 1 mm2 areas of peritumoral stroma were counted by an observer unaware of the treatments using a computer-assisted image analysis system, which included a Nikon Eclipse E600 microscope equipped with a DS-5M digital color video camera. Images were captured through a PC interface using the software package Lucia G 4.81 (Laboratory Imaging Ltd., Praha, Hostivar). The number of immunoreactive cells in each group was then calculated by averaging the cell numbers from all specimens. The mean and SEM were calculated for each parameter. Two-way ANOVA was used to assess the treatment effects, and one-way analysis of variance and Tukey’s multiple comparison post hoc test were used to determine differences among groups. P < 0.05 served as the level for statistical significance.
Immunohistochemistry in human tissue specimens
A total of 39 patients with lung carcinoma (18 patients) or melanoma (21 patients) who underwent surgical resection at the University Hospital of Ferrara were included in the study. Their clinical features are reported in Table S1; for all of the cases, histopathological parameter analysis was performed by pathologists of the Pathology Unit of the University Hospital of Ferrara. The study was approved by the Local Ethics Committee (CE: 1016/2020/Oss/UniFe). The informed consent was collected from patients whenever it was possible. CD68 immunodetection was performed on formalin-fixed paraffin-embedded tissue sections (4 µm thick) using the antibody CD68 (PG-M1, Diagnostic Biosystems) and the Multimeric Detection Kit (Universal DAB Detection Kit Ultraview, Roche Tissue Diagnostics (CH)) were assessed on a Benchmark XT automated immunostainer (Roche T. D.). For the immunohistochemical evaluation of PML, IL-1β and CD206, 4 µm-thick tissue sections were immunostained with anti-PML (Abcam ab72137, 1:250 dilution), anti-IL-1β (Αbcam ab9722, 1:500 dilution) and anti-CD206 (Abcam ab64693, 1:1000 dilution). After deparaffinization, immunohistochemistry was performed according to [38].
The immunoreactivities of CD68-, PML-, IL-1β−, and CD206 in the peritumoral stroma were evaluated by two independent clinical pathologists (blinded to the patient outcomes) and scored for occurrence/absence.
Immunofluorescence assay
pRMØs from WT and pml−/− mice were seeded on 24-mm glass coverslips; the next day, the O/N medium was replaced, and cells were stimulated with LPS (1 µg/ml for 2 h) plus nigericin (10 µM for 1 h). Then, the cells were fixed with 4% paraformaldehyde (PFA) in PBS for 10 min and washed three times with PBS. The cells were permeabilized for 10 min with 0.1% Triton X-100 in PBS and blocked with PBS containing 2% BSA and 0.1% Triton X-100 for 1 h. The cells were incubated O/N at 4 °C with the following antibodies: calnexin (Santa Cruz Biotechnology, sc11397) and NLRP3 (Adipogen, AG-20B-0014-C100). Appropriate isotype-matched AlexaFluor-conjugated secondary antibodies (diluted 1:1000) were then used (Life Technologies A11008 488 goat anti-rabbit and A-11005 goat anti-mouse 594). The coverslips were mounted with ProLong Gold Antifade reagent (Life Technologies), and immunofluorescence analysis was performed with a confocal laser scanner microscope (Olympus FV3000) equipped with a 63x oil objective. The NLRP3 speckle/NLRP3 total ratio was quantified.
Co-immunoprecipitation
Co-immunoprecipitation was carried out using protein Gibco CTS Dynabeads (Thermo Fisher) in accordance with the manufacturer’s instructions. Protein extractions from the ER fractions were carried out by adding 50 mM NaCl and 1% NP-40 to the homogenization buffer. All the buffers were supplemented with a proteases inhibitor mixture and Phos‐STOP Phosphatase Inhibitor Cocktail (Roche Applied Science). The same amount of extracted proteins for each condition was incubated O/N with the specific primary antibody PML (Millipore, no. MAB3738), P2X7 (Alomone, APR-004), or NLRP3 (AG-20B-0014-C100, Adipogen). The immunocomplexes were captured with the appropriate dynabeads (Thermo Fisher). Beads were pelleted and washed three times. The bait was eluted in Laemmli sample buffer and denatured for 5 min at 100 °C. Samples were proceeded by SDS–PAGE and analyzed by standard (WB) blotting technique.
RT-PCR
Total RNA was extracted with TRIzol reagent (Life Technologies) and then purified with the Pure Link RNA Mini Kit (Invitrogen) in accordance with the manufacturer’s instructions. The RNA was quantified using a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, Milan, Italy). One microgram of the total RNA extract was reverse transcribed into cDNA using the High Capacity cDNA Reverse Transcription kit (Applied Biosystem, Carlsbad, CA, USA). qRT PCR was performed on a Step One Real-Time PCR System (Applied Biosystems). Two microliters of cDNA were used as a template. Amplification was performed using predesigned TaqMan probes (Applied Biosystems) for NLRP3, NLRC4 and GAPDH as housekeeping genes. For the screening of CRISPR/Cas9 KO clones, RT–PCR was performed with Two-Step Real-Time PCR on the Rotor Gene Q system (Qiagen). Two microliters of cDNA were used as a template. Amplification of IL-1R1 and GAPDH, a housekeeping gene, was performed using predesigned primers (IDTs).
PCR
DNA was prepared from tail biopsies and used in 50 μl reaction mixtures that included the following specific primers for each selective gene being amplified: (I) 5′-TGCCCATCTTCTGAACACC, 5′-CTTCCTCTTACTGTTTCCTCCC, and 5′-GCAAGGCGATTAAGTTGGG for P2X7R; (II) 5′-GTCCAGGACATACGTCTGGA, 5′TGAGGTCCACATCTTCAAGG and 5′TTGTAGTTGCCGTCGTCCTT for NLRP3; and (III) 5′-TTGGACTTGCGCGTACTGTC, 5′-CGACCACCAAGCGAAACA, and 5′-TTTCAGTTTCTGCGCTGCC for PML.
WT genes corresponded to specific amplicons (568 bp for P2x7r; 327 bp for Nlrp3; 400 bp for Pml), unlike the respective KO gene amplicons (393 bp for P2x7r−/−; 589 bp for Nlrp3−/−; 700 bp for Pml−/−). The reactions were analyzed on a 2% agarose gel containing Midori green.
Flow cytometry
Tumors were cut into 1–2 mm3 pieces and digested with DNAase (100 μg/ml, Sigma) and collagenase A (20 units/ml, Roche) in RPMI at 37 °C for 1 h. TAMs were blocked with an anti-mouse FcR antibody (CD16/CD32, BD) for 5 min at 4 °C; subsequently, the cells were surface stained in the dark for 15 min at room temperature with directly conjugated monoclonal antibodies (mAbs) and analyzed after the exclusion of nonviable cells using the amine-reactive viability dye Aqua (Life Technologies) as previously described (1, 2). The following mAbs were used to identify macrophages (see Extended Data Fig. 4b for the gating strategy) and analyze their polarization: APC-conjugated anti-CD3 (clone: 145-2C11, eBioscience), APC-conjugated anti-CD45R (clone: RA3-6B2, eBioscience), PE/Cy7-conjugated anti-CD11b (clone: M1/70, Biolegend), APC/Cy7-conjugated anti-F4/80 (clone: BM8, Biolegend), PE-conjugated anti-CD86 (clone: PO3, Biolegend), and Pacific Blue-conjugated anti-MHC-II (clone: M5/114.15.2, Biolegend). To determine the percentage of circulating monocytes, samples of whole blood were immunostained using the following mAbs: PE-conjugated anti-45 (clone: 30F11, Miltenyi), APC-conjugated anti-CD11c (clone: N418, Biolegend), PE/Cy7-conjugated anti-CD11b (clone: M1/70, Biolegend), PerCP-vio700-conjugated anti-Ly-6C (clone REA796, Miltenyi), and VioBlue-conjugated anti-Ly-6G (clone: REA526, Miltenyi). Samples were acquired on a BD FACSCantoII (BD Biosciences) flow cytometer. Data were analyzed using FlowJo version 10 (Tree Star Inc.).
Bone marrow replacement experiment
B6. SJL-Ptprca Pepcb/BoyJ (Jackson Laboratory) recipients received 950 cGy of total body irradiation, followed by the intravenous injection of 106 bone marrow mononuclear cells derived from WT or pml−/− C57BL/6J donor mice.
Reconstitution of donor cells and repopulation of donor myeloid and lymphoid cells were monitored by the staining of peripheral blood cells with antibodies against CD45.1, CD45.2, CD3 (T cell), B220 (B cell), CD11b and Gr-1 (myeloid) every 4 weeks starting at 4 weeks after transplantation. At 16 weeks post transplantation, the engraftments were considered stable, and the mice were subjected to tumor engraftment. Experiments were authorized by the Institutional Review Board for Human Research at Albert Einstein College of Medicine.
Surface plasmon resonance (SPR) experiments
Recombinant P2X7 protein (GST tag; Cat. No. ABIN1313816; Fc tag; Cat. No. ABIN16964308), NLRP3 protein (AA 1-1036; GST tag; Cat. No. ABIN1312675) and PML protein (Transcript Variant 9, Myc-DYKDDDDK tag; Cat. No. ABIN2729208) were purchased from Antibodies Online, Germany. SPR measurements were performed on a Biacore 3000 instrument (GE Healthcare/Cytiva). GST-tagged NLRP3 protein was captured at low surface density on a polyclonal goat anti-GST antibody, covalently coupled to the surface of a Biacore CM5 optical sensor chip, using and following the protocol of the Biacore GST Capture Kit (Order Code: BR-1002-23). Amine activated flow cell 1 (FC1) of the sensor chip was used as a reference to allow the generation of background-subtracted binding sensorgrams. After GST-tagged NLRP3 or P2RX7 ligand capturing, remaining free anti-GST binding sites were saturated with excess recombinant GST provided with the Biacore GST Capture Kit to allow specific sequential binding of other GST-tagged analytes to the captured ligand protein. Proteins PML and P2X7R were passed as analytes in sequential injections over the captured NLRP3 ligand at single concentrations of 167 nM at a flow rate 30 μl/minutes in HBS-EP buffer (0.1 M HEPES, 1.5 M NaCl, 0.03 M EDTA and 0.5% v/v Surfactant P20). PML was also passed over captured P2X7R in a separate experiment. After the experiments bound captured ligand and analyte proteins were removed by anti-GST surface regeneration with 10 mM HCl.
Proximity ligation assay
pRMØs from WT and pml−/− mice were seeded on 16-well microarray slides (Thermo Fisher) at a density of 105 cells. Twenty-four hours after seeding, the cells were primed with LPS (1 µg/ml for 2 h) and then with nigericin (10 µM for 1 h). The cells were then fixed with 4% PFA in PBS for 10 min and washed three times with PBS. The cells were permeabilized for 10 min with 0.1% Triton X-100 in PBS and blocked with PBS containing 5% BSA and 0.1% Triton X-100 for 1 h. Then, the cells were hybridized (O/N at 4 °C in a humid chamber) with the following conjugated primary antibodies (PLA probes) overnight: one MINUS (P2X7R) and one PLUS (NLRP3) for the P2X7R-NLRP3 interaction, one MINUS (PML) and one PLUS (NLRP3) for the PML-NLRP3 interaction, and one MINUS (P2X7R) and one PLUS (PML) for the P2X7R-PML interaction. The next day, the PLA was conducted in accordance with the manufacturer’s instructions. After the PLA assay, the cells were fixed with 100% methanol for 20 min at −20 °C and then permeabilized for 10 min with 0.1% Triton X-100 in PBS. The cells were blocked with PBS containing 5% milk and 0.1% Triton X-100 for 1 h and then incubated O/N at 4 °C with an antibody against PDI (Abcam, ab3672). An appropriate, isotype-matched AlexaFluor-conjugated secondary antibody (Life Technologies, A11008, 488 goat anti-rabbit, diluted 1:1000) was then applied, and the PLA signal was detected by an Olympus Xcellence widefield system and deconvolved using Fiji. After 3D digital deconvolution, the PLA signal was quantified as dots within each PDI-positive cell or DAPI-positive cell.
Statistics analyses
The data were analyzed by Prism 6 (GraphPad) or Microsoft Excel (Microsoft Co.). Unless otherwise specified, the data are representative of at least three biologically independent experiments. Two-group datasets were analyzed by Student’s t test for unpaired data. For multiple comparison (≥3 experimental groups) analysis, one-way ANOVA followed by Tukey’s or Dunnett’s test was used where appropriate. A P value <0.05 was considered significant.
Data availability
The data analyzed during this study are included in this article and the supplemental data files. Additional supporting data are available from the corresponding authors upon reasonable request.
References
Pinton P, Giorgi C, Pandolfi PP. The role of PML in the control of apoptotic cell fate: a new key player at ER-mitochondria sites. Cell Death Differ. 2011;18:1450–6.
Giorgi C, Ito K, Lin HK, Santangelo C, Wieckowski MR, Lebiedzinska M, et al. PML regulates apoptosis at endoplasmic reticulum by modulating calcium release. Science. 2010;330:1247–51.
Missiroli S, Bonora M, Patergnani S, Poletti F, Perrone M, Gafa R, et al. PML at mitochondria-associated membranes is critical for the repression of autophagy and cancer development. Cell Rep. 2016;16:2415–27.
Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–5.
Missiroli S, Patergnani S, Caroccia N, Pedriali G, Perrone M, Previati M, et al. Mitochondria-associated membranes (MAMs) and inflammation. Cell Death Dis. 2018;9:329.
Karki R, Kanneganti TD. Diverging inflammasome signals in tumorigenesis and potential targeting. Nat Rev Cancer. 2019;19:197–214.
Latz E, Xiao TS, Stutz A. Activation and regulation of the inflammasomes. Nat Rev Immunol. 2013;13:397–411.
Karmakar M, Katsnelson MA, Dubyak GR, Pearlman E. Neutrophil P2X7 receptors mediate NLRP3 inflammasome-dependent IL-1beta secretion in response to ATP. Nat Commun. 2016;7:10555.
Di Virgilio F, Dal Ben D, Sarti AC, Giuliani AL, Falzoni S. The P2X7 receptor in infection and inflammation. Immunity. 2017;47:15–31.
Franceschini A, Capece M, Chiozzi P, Falzoni S, Sanz JM, Sarti AC, et al. The P2X7 receptor directly interacts with the NLRP3 inflammasome scaffold protein. FASEB J. 2015;29:2450–61.
Adinolfi E, Giuliani AL, De Marchi E, Pegoraro A, Orioli E, Di Virgilio F. The P2X7 receptor: a main player in inflammation. Biochem Pharmacol. 2018;151:234–44.
Kelley N, Jeltema D, Duan Y, He Y. The NLRP3 inflammasome: an overview of mechanisms of activation and regulation. Int J Mol Sci. 2019;20:3328.
Tweedell RE, Malireddi RKS, Kanneganti TD. A comprehensive guide to studying inflammasome activation and cell death. Nat Protoc. 2020;15:3284–333.
Coll RC, Robertson AA, Chae JJ, Higgins SC, Munoz-Planillo R, Inserra MC, et al. A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat Med. 2015;21:248–55.
Adinolfi E, Raffaghello L, Giuliani AL, Cavazzini L, Capece M, Chiozzi P, et al. Expression of P2X7 receptor increases in vivo tumor growth. Cancer Res. 2012;72:2957–69.
Broz P, Dixit VM. Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol. 2016;16:407–20.
Missiroli S, Perrone M, Boncompagni C, Borghi C, Campagnaro A, Marchetti F, et al. Targeting the NLRP3 inflammasome as a new therapeutic option for overcoming cancer. Cancers. 2021;13:2297.
Mantovani A, Barajon I, Garlanda C. IL-1 and IL-1 regulatory pathways in cancer progression and therapy. Immunol Rev. 2018;281:57–61.
Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–99.
Guerriero JL. Macrophages: the road less traveled, changing anticancer therapy. Trends Mol Med. 2018;24:472–89.
Lin Y, Xu J, Lan H. Tumor-associated macrophages in tumor metastasis: biological roles and clinical therapeutic applications. J Hematol Oncol. 2019;12:76.
Arwert EN, Harney AS, Entenberg D, Wang Y, Sahai E, Pollard JW, et al. A unidirectional transition from migratory to perivascular macrophage is required for tumor cell intravasation. Cell Rep. 2018;23:1239–48.
Franklin RA, Li MO. Ontogeny of tumor-associated macrophages and its implication in cancer regulation. Trends Cancer. 2016;2:20–34.
Lo YH, Huang YW, Wu YH, Tsai CS, Lin YC, Mo ST, et al. Selective inhibition of the NLRP3 inflammasome by targeting to promyelocytic leukemia protein in mouse and human. Blood. 2013;121:3185–94.
Dowling JK, Becker CE, Bourke NM, Corr SC, Connolly DJ, Quinn SR, et al. Promyelocytic leukemia protein interacts with the apoptosis-associated speck-like protein to limit inflammasome activation. J Biol Chem. 2014;289:6429–37.
Liu CY, Xu JY, Shi XY, Huang W, Ruan TY, Xie P, et al. M2-polarized tumor-associated macrophages promoted epithelial-mesenchymal transition in pancreatic cancer cells, partially through TLR4/IL-10 signaling pathway. Lab Investig. 2013;93:844–54.
Zhang Q, Wang H, Mao C, Sun M, Dominah G, Chen L, et al. Fatty acid oxidation contributes to IL-1beta secretion in M2 macrophages and promotes macrophage-mediated tumor cell migration. Mol Immunol. 2018;94:27–35.
Wang YT, Chen J, Chang CW, Jen J, Huang TY, Chen CM, et al. Ubiquitination of tumor suppressor PML regulates prometastatic and immunosuppressive tumor microenvironment. J Clin Investig. 2017;127:2982–97.
Scherer M, Stamminger T. Emerging role of PML nuclear bodies in innate immune signaling. J Virol. 2016;90:5850–4.
Lin S, Mei X. Role of NLRP3 inflammasomes in neuroinflammation diseases. Eur Neurol. 2020;83:576–80.
Kuchay S, Giorgi C, Simoneschi D, Pagan J, Missiroli S, Saraf A, et al. PTEN counteracts FBXL2 to promote IP3R3- and Ca(2+)-mediated apoptosis limiting tumour growth. Nature. 2017;546:554–8.
Ying W, Cheruku PS, Bazer FW, Safe SH, Zhou B. Investigation of macrophage polarization using bone marrow derived macrophages. J Vis Exp. 2013;50323.
Zhang X, Goncalves R, Mosser DM. The isolation and characterization of murine macrophages. Curr Protoc Immunol. 2008;Chapter 14:Unit 14.1.
Riedhammer C, Halbritter D, Weissert R. Peripheral blood mononuclear cells: isolation, freezing, thawing, and culture. Methods Mol Biol. 2016;1304:53–61.
Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–308.
Vance JE. Phospholipid synthesis in a membrane fraction associated with mitochondria. J Biol Chem. 1990;265:7248–56.
Wieckowski MR, Giorgi C, Lebiedzinska M, Duszynski J, Pinton P. Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nat Protoc. 2009;4:1582–90.
Marchi S, Corricelli M, Trapani E, Bravi L, Pittaro A, Delle Monache S, et al. Defective autophagy is a key feature of cerebral cavernous malformations. EMBO Mol Med. 2015;7:1403–17.
Acknowledgements
The authors thank C. Bosi, F. Poletti, A.C. Sarti, and S. Falzoni for their contribution to this study. PP is grateful to C. degli Scrovegni for her continuous support.
Funding
The Signal Transduction Laboratory is supported by the Italian Association for Cancer Research (IG-23670 to PP, IG-19803 to CG and IG 22883 to FDV), A-ROSE, Progetti di Rilevante Interesse Nazionale (PRIN2017E5L5P3 to PP and PRIN20177E9EPY to CG), the Italian Ministry of Health (GR-2019-12369646 to SM), the European Research Council (853057-InflaPML to CG) and local funds from the University of Ferrara to CG and PP. MRW was supported by the National Science Centre, Poland (UMO-2018/29/B/NZ1/00589). MLA was funded by a Polish National Science Centre grant (UMO-2015/17/D/NZ1/00030). MP is supported by an AIRC research fellowship (ID: 26665).
Author information
Authors and Affiliations
Contributions
CG conceived the study. SM performed most of the experiments and prepared the figures. MP and SMa performed the proximity ligation assay studies. MP generated the CRISPR knockout clones. CG directed and coordinated the study, oversaw all the experiments, and wrote the manuscript. PP supervised the study and oversaw the experiments. F.D.V. collaborated in experiments related to P2X7R. GM, MRW, and MLA performed the co-immunoprecipitation experiments. RG, GL and LA performed the immunohistochemical analysis. LA helped in writing the manuscript. SM and MP provided advice for the in vivo experiments. FN performed the flow cytometry experiments. MB and KI performed the bone marrow replacement experiment. CB and BV helped with some experiments. SM performed the Western blot, ELISA, and subcellular fractionation studies. FK performed the SPR studies. FF, AB, and RG helped in the patient’s cohorts selection. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
Procedures involving animals and their care were in conformity with institutional guidelines, and the Animal Ethics Committee approved all experimental protocols (authorization nos. 481/2017-PR and CBCC2.N.BH4 approved by the Italian Ministry of Health). The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Ethics Committee of the University Hospital of Ferrara (CE-AVEC: 1016/2020/Oss/UniFe).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by P Salomoni
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Missiroli, S., Perrone, M., Gafà, R. et al. PML at mitochondria-associated membranes governs a trimeric complex with NLRP3 and P2X7R that modulates the tumor immune microenvironment. Cell Death Differ 30, 429–441 (2023). https://doi.org/10.1038/s41418-022-01095-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-022-01095-9