Abstract
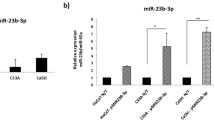
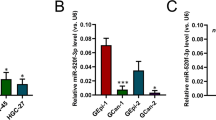
Although significant advances have recently been made in the diagnosis and treatment of cervical carcinoma, the long-term survival rate for advanced cervical cancer remains low. Therefore, an urgent need exists to both uncover the molecular mechanisms and identify potential therapeutic targets for the treatment of cervical cancer. MicroRNAs (miRNAs) have important roles in cancer progression and could be used as either potential therapeutic agents or targets. miR-506 is a component of an X chromosome-linked miRNA cluster. The biological functions of miR-506 have not been well established. In this study, we found that miR-506 expression was downregulated in approximately 80% of the cervical cancer samples examined and inversely correlated with the expression of Ki-67, a marker of cell proliferation. Gain-of-function and loss-of-function studies in human cervical cancer, Caski and SiHa cells, demonstrated that miR-506 acts as a tumor suppressor by inhibiting cervical cancer growth in vitro and in vivo. Further studies showed that miR-506 induced cell cycle arrest at the G1/S transition, and enhanced apoptosis and chemosensitivity of cervical cancer cell. We subsequently identified Gli3, a hedgehog pathway transcription factor, as a direct target of miR-506 in cervical cancer. Furthermore, Gli3 silencing recapitulated the effects of miR-506, and reintroduction of Gli3 abrogated miR-506-induced cell growth arrest and apoptosis. Taken together, we conclude that miR-506 exerts its anti-proliferative function by directly targeting Gli3. This newly identified miR-506/Gli3 axis provides further insight into the pathogenesis of cervical cancer and indicates a potential novel therapeutic agent for the treatment of cervical cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D . Global cancer statistics. CA Cancer J Clin 2011; 61: 69–90.
Chemoradiotherapy for Cervical Cancer Meta-Analysis Collaboration, Reducing uncertainties about the effects of chemoradiotherapy for cervical cancer: a systematic review and meta-analysis of individual patient data from 18 randomized trials. J Clin Oncol 2008; 26: 5802–5812.
Ambros V, Lee RC . Identification of microRNAs and other tiny noncoding RNAs by cDNA cloning. Methods Mol Biol 2004; 265: 131–158.
McManus MT . MicroRNAs and cancer. Sem Cancer Biol 2003; 13: 253–258.
Lim YY, Wright JA, Attema JL, Gregory PA, Bert AG, Smith E et al. Epigenetic modulation of the miR-200 family is associated with transition to a breast cancer stem-cell-like state. J Cell Sci 2013; 126: 2256–2266.
Mitra AK, Zillhardt M, Hua Y, Tiwari P, Murmann AE, Peter ME et al. MicroRNAs reprogram normal fibroblasts into cancer-associated fibroblasts in ovarian cancer. Cancer Discov 2012; 2: 1100–1108.
Sun MM, Li JF, Guo LL, Xiao HT, Dong L, Wang F et al. TGF-β1 suppression of microRNA-450b-5p expression: a novel mechanism for blocking myogenic differentiation of rhabdomyosarcoma. Oncogene (e-pub ahead of print 13 May 2013; doi:10.1038/onc.2013.165).
Roy SS, Gonugunta VK, Bandyopadhyay A, Rao MK, Goodall GJ, Sun L-Z et al. Significance of PELP1/HDAC2/miR-200 regulatory network in EMT and metastasis of breast cancer. Oncogene (e-pub ahead of print 26 August 2013; doi:10.1038/onc.2013.332).
Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet 2005; 37: 766–770.
Castro RE, Rodrigues CM . Targeting miR-506 in primary biliary cirrhosis to support the HCO3- umbrella. Clin Res Hepatol Gastroenterol 2012; 36: 402–404.
Banales JM, Saez E, Uriz M, Sarvide S, Urribarri AD, Splinter P et al. Up-regulation of microRNA 506 leads to decreased Cl-/HCO3- anion exchanger 2 expression in biliary epithelium of patients with primary biliary cirrhosis. Hepatology 2012; 56: 687–697.
Streicher KL, Zhu W, Lehmann KP, Georgantas RW, Morehouse CA, Brohawn P et al. A novel oncogenic role for the miRNA-506-514 cluster in initiating melanocyte transformation and promoting melanoma growth. Oncogene 2011; 31: 1558–1570.
Tong JL, Zhang CP, Nie F, Xu XT, Zhu MM, Xiao SD et al. MicroRNA 506 regulates expression of PPAR alpha in hydroxycamptothecin-resistant human colon cancer cells. FEBS Lett 2011; 585: 3560–3568.
Yang D, Sun Y, Hu L, Zheng H, Ji P, Pecot Chad V et al. Integrated Analyses Identify a Master MicroRNA Regulatory Network for the Mesenchymal Subtype in Serous Ovarian Cancer. Cancer Cell 2013; 23: 186–199.
Zhao Y, Liu H, Li Y, Wu J, Greenlee AR, Yang C et al. The role of miR-506 in transformed 16HBE cells induced by anti-benzo[a]pyrene-trans-7,8-dihydrodiol-9,10-epoxide. Toxicol Lett 2011; 205: 320–326.
Kang HN, Oh SC, Kim JS, Yoo YA . Abrogation of Gli3 expression suppresses the growth of colon cancer cells via activation of p53. Exp Cell Res 2012; 318: 539–549.
Steg A, Amm HM, Novak Z, Frost AR, Johnson MR . Gli3 mediates cell survival and sensitivity to cyclopamine in pancreatic cancer. Cancer Biol Ther 2010; 10: 893–902.
Kim KH, Kim JM, Choi Y-L, Shin YK, Lee H-c, Seong IO et al. Expression of Sonic hedgehog signaling molecules in normal, hyperplastic and carcinomatous endometrium. Pathol Int 2009; 59: 279–287.
Liu K, Zhao H, Yao H, Lei S, Lei Z, Li T et al. MicroRNA-124 regulates the proliferation of colorectal cancer cells by targeting iASPP. Biomed Res Int 2013; 2013: 1–10.
Wang P, Chen L, Zhang J, Chen H, Fan J, Wang K et al. Methylation-mediated silencing of the miR-124 genes facilitates pancreatic cancer progression and metastasis by targeting Rac1. Oncogene 2013; 33: 514–524.
Furuta M, Kozaki Ki, Tanaka S, Arii S, Imoto I, Inazawa J . miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis 2009; 31: 766–776.
Silber J, Lim DA, Petritsch C, Persson AI, Maunakea AK, Yu M et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med 2008; 6: 14.
Pierson J, Hostager B, Fan R, Vibhakar R . Regulation of cyclin dependent kinase 6 by microRNA 124 in medulloblastoma. J Neurooncol 2008; 90: 1–7.
Li KKW, Pang JC-s, Ching, Wong CK, Kong X, Wang Y et al. miR-124 is frequently down-regulated in medulloblastoma and is a negative regulator of SLC16A1. Hum Pathol 2009; 40: 1234–1243.
Shi XB, Xue L, Ma AH, Tepper CG, Gandour-Edwards R, Kung HJ et al. Tumor suppressive miR-124 targets androgen receptor and inhibits proliferation of prostate cancer cells. Oncogene 2012; 32: 4130–4138.
Wilting SM, van Boerdonk RAA, Henken FE, Meijer CJLM, Diosdado B, Meijer GA et al. Methylation-mediated silencing and tumour suppressive function of hsa-miR-124 in cervical cancer. Mol Cancer 2010; 9: 167.
Renault MA, Roncalli J, Tongers J, Misener S, Thorne T, Jujo K et al. The hedgehog transcription factor gli3 modulates angiogenesis. Circ Res 2009; 105: 818–826.
Sengupta A, Banerjee D, Chandra S, Banerji SK, Ghosh R, Roy R et al. Deregulation and cross talk among Sonic hedgehog, Wnt, Hox and Notch signaling in chronic myeloid leukemia progression. Leukemia 2007; 21: 949–955.
Iwasaki H, Nakano K, Shinkai K, Kunisawa Y, Hirahashi M, Oda Y et al. Hedgehog Gli3 activator signal augments tumorigenicity of colorectal cancer via upregulation of adherence-related genes. Cancer Sci 2013; 104: 328–336.
Renault MA, Roncalli J, Tongers J, Misener S, Thorne T, Jujo K et al. The hedgehog transcription factor Gli3 modulates angiogenesis. Circ Res 2009; 105: 818–826.
Scherr M, Venturini L, Battmer K, Schaller-Schoenitz M, Schaefer D, Dallmann I et al. Lentivirus-mediated antagomir expression for specific inhibition of miRNA function. Nucleic Acids Res 2007; 35: e149–e149.
Kutner RH, Zhang X-Y, Reiser J . Production, concentration and titration of pseudotyped HIV-1-based lentiviral vectors. Nat Protocols 2009; 4: 495–505.
He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y et al. A microRNA component of the p53 tumour suppressor network. Nature 2007; 447: 1130–1134.
Acknowledgements
We are most grateful for Dr Lei-Zhu, Bo-Shi Wang, Ming-Xuan Feng and Doctor XingLin Yang of Medical and Biological Engineering Technology Co. Ltd of Heyuan of Shanghai. This work was supported by the National Science Foundation of China (no. 81071738; no.81101600; no. 81201624), Songjiang district of Science and Technology Commission of Shanghai Municipality (no.10SJGG26), and Shanghai Songjiang District Central Hospital (no. BY10A07).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Rights and permissions
About this article
Cite this article
Wen, SY., Lin, Y., Yu, YQ. et al. miR-506 acts as a tumor suppressor by directly targeting the hedgehog pathway transcription factor Gli3 in human cervical cancer. Oncogene 34, 717–725 (2015). https://doi.org/10.1038/onc.2014.9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2014.9
This article is cited by
-
Study of MicroRNA Cluster Located on Chromosome X in Serum and Breast Cancer Tissue
Biochemical Genetics (2023)
-
Inhibition of CD4 + T cells by fanchinoline via miR506-3p/NFATc1 in Sjögren’s syndrome
Inflammopharmacology (2023)
-
Turing miRNA into infinite coordination supermolecule: a general and enabling nanoengineering strategy for resurrecting nuclear acid therapeutics
Journal of Nanobiotechnology (2022)
-
DSCAM-AS1 promotes cervical carcinoma cell proliferation and invasion via sponging miR-338-3p
Environmental Science and Pollution Research (2022)
-
ChrXq27.3 miRNA cluster functions in cancer development
Journal of Experimental & Clinical Cancer Research (2021)