Abstract
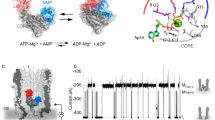
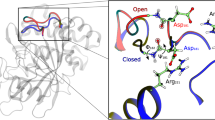
A fundamental question is how enzymes can accelerate chemical reactions. Catalysis is not only defined by actual chemical steps, but also by enzyme structure and dynamics. To investigate the role of protein dynamics in enzymatic turnover, we measured residue-specific protein dynamics in hyperthermophilic and mesophilic homologs of adenylate kinase during catalysis. A dynamic process, the opening of the nucleotide-binding lids, was found to be rate-limiting for both enzymes as measured by NMR relaxation. Moreover, we found that the reduced catalytic activity of the hyperthermophilic enzyme at ambient temperatures is caused solely by a slower lid-opening rate. This comparative and quantitative study of activity, structure and dynamics revealed a close link between protein dynamics and catalytic turnover.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Jaenicke, R. & Bohm, G. The stability of proteins in extreme environments. Curr. Opin. Struct. Biol. 8, 738–748 (1998).
Petsko, G.A. Structural basis of thermostability in hyperthermophilic proteins, or “there's more than one way to skin a cat.” Methods Enzymol. 334, 469–478 (2001).
Zavodszky, P., Kardos, J., Svingor & Petsko, G.A. Adjustment of conformational flexibility is a key event in the thermal adaptation of proteins. Proc. Natl. Acad. Sci. USA 95, 7406–7411 (1998).
D'Amico, S., Marx, J.C., Gerday, C. & Feller, G. Activity-stability relationships in extremophilic enzymes. J. Biol. Chem. 278, 7891–7896 (2003).
Kohen, A., Cannio, R., Bartolucci, S. & Klinman, J.P. Enzyme dynamics and hydrogen tunnelling in a thermophilic alcohol dehydrogenase. Nature 399, 496–499 (1999).
Vonrhein, C., Schlauderer, G.J. & Schulz, G.E. Movie of the structural changes during a catalytic cycle of nucleoside monophosphate kinases. Structure 3, 483–490 (1995).
Rhoads, D.G. & Lowenstein, J.M. Initial velocity and equilibrium kinetics of myokinase. J. Biol. Chem. 243, 3963–3972 (1968).
Loria, J.P., Rance, M. & Palmer, A.G. A relaxation-compensated Carr-Purcell-Meiboom-Gill sequence for characterizing chemical exchange by NMR spectroscopy. J. Am. Chem. Soc. 121, 2331–2332 (1999).
Tollinger, M., Skrynnikov, N.R., Mulder, F.A., Forman-Kay, J.D. & Kay, L.E. Slow dynamics in folded and unfolded states of an SH3 domain. J. Am. Chem. Soc. 123, 11341–11352 (2001).
Palmer, A.G., Kroenke, C.D. & Loria, J.P. Nuclear magnetic resonance methods for quantifying microsecond-to-millisecond motions in biological macromolecules. Methods Enzymol. 339, 204–238 (2001).
Bruice, T.C. & Benkovic, S.J. Chemical basis for enzyme catalysis. Biochemistry 39, 6267–6274 (2000).
Jencks, W.P. Catalysis in Chemistry and Biology (Dover, New York, 1987).
Mulder, F.A., Mittermaier, A., Hon, B., Dahlquist, F.W. & Kay, L.E. Studying excited states of proteins by NMR spectroscopy. Nat. Struct. Biol. 8, 932–935 (2001).
Muller, C.W. & Schulz, G.E. Structure of the complex between adenylate kinase from Escherichia coli and the inhibitor Ap5A refined at 1.9 Å resolution. A model for a catalytic transition state. J. Mol. Biol. 224, 159–177 (1992).
Muller, C.W., Schlauderer, G.J., Reinstein, J. & Schulz, G.E. Adenylate kinase motions during catalysis: an energetic counterweight balancing substrate binding. Structure 4, 147–156 (1996).
Berry, M.B. et al. The closed conformation of a highly flexible protein: the structure of E. coli adenylate kinase with bound AMP and AMPPNP. Proteins 19, 183–198 (1994).
Rozovsky, S., Jogl, G., Tong, L. & McDermott, A.E. Solution-state NMR investigations of triosephosphate isomerase active site loop motion: ligand release in relation to active site loop dynamics. J. Mol. Biol. 310, 271–280 (2001).
Osborne, M.J., Schnell, J., Benkovic, S.J., Dyson, H.J. & Wright, P.E. Backbone dynamics in dihydrofolate reductase complexes: role of loop flexibility in the catalytic mechanism. Biochemistry 40, 9846–9859 (2001).
Nicholson, L.K. et al. Flexibility and function in HIV-1 protease. Nat. Struct. Biol. 2, 274–280 (1995).
Wang, L. et al. Functional dynamics in the active site of the ribonuclease binase. Proc. Natl. Acad. Sci. USA 98, 7684–7689 (2001).
Sinev, M.A., Sineva, E.V., Ittah, V. & Haas, E. Domain closure in adenylate kinase. Biochemistry 35, 6425–6437 (1996).
Reinstein, J., Brune, M. & Wittinghofer, A. Mutations in the nucleotide binding loop of adenylate kinase of Escherichia coli. Biochemistry 27, 4712–4720 (1988).
Burlacu-Miron, S. et al. 1H, 13C and 15N backbone resonance assignment of Escherichia coli adenylate kinase, a 23.6 kDa protein. J. Biomol. NMR 13, 93–94 (1999).
Cavanagh, J., Fairbrother, W.J., Palmer, A.G. & Skelton, N.J. Protein NMR Spectroscopy Principles and Practice (Academic Press, San Diego, 1996).
Carver, J.P. & Richards, R.E. A general two-site solution for the chemical exchange produced dependence of T2 upon the Carr-Purcell pulse separation. J. Magn. Reson. 6, 89–105 (1972).
Millet, O., Loria, J.P., Kroenke, C.D., Pons, M. & Palmer, A.G. The static magnetic field dependence of chemical exchange linebroadening defines the NMR chemical shift time scale. J. Am. Chem. Soc. 122, 2867–2877 (2000).
Binsch, G. A unified theory of exchange effects on nuclear magnetic resonance line shapes. J. Am. Chem. Soc. 91, 1304–1309 (1969).
Koradi, R., Billeter, M. & Wuthrich, K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14, 51–55, 29–32 (1996).
Acknowledgements
We thank The Swedish NMR Centre for 800 MHz NMR spectrometer time, V. Orekhov for valuable technical advice on the acquisition of NMR relaxation dispersion experiments, L. Kay for providing pulse programs and D. Korzhnev for sharing software for NMR relaxation data analysis. We are grateful to A. Wittinghofer for supplying the plasmid for E. coli adenylate kinase and K.O. Stetter for providing DNA isolated from Aquifex aeolicus. This work was supported by US National Institutes of Health (NIH) grants GM067963 and GM62117 to D.K. and a postdoctoral fellowship from the Swedish Research Council to M.W.W. and from the NIH to K.H.W.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Wolf-Watz, M., Thai, V., Henzler-Wildman, K. et al. Linkage between dynamics and catalysis in a thermophilic-mesophilic enzyme pair. Nat Struct Mol Biol 11, 945–949 (2004). https://doi.org/10.1038/nsmb821
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb821