Abstract
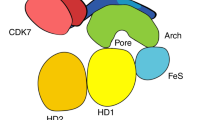
The human general transcription factor TFIIH is involved in both transcription and DNA repair. We have identified a structural domain in the core subunit of TFIIH, p62, which is absolutely required for DNA repair activity through the nucleotide excision repair pathway. Using coimmunoprecipitation experiments, we showed that this activity involves the interaction between the N-terminal domain of p62 and the 3′ endonuclease XPG, a major component of the nucleotide excision repair machinery. Furthermore, we reconstituted a functional TFIIH particle with a mutant of p62 lacking the N-terminal domain, showing that this domain is not required for assembly of the TFIIH complex and basal transcription. We solved its three-dimensional structure and found an unpredicted pleckstrin homology and phosphotyrosine binding (PH/PTB) domain, uncovering a new class of activity for this fold.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Coin, F. & Egly, J.M. Ten years of TFIIH. Cold Spring Harb. Symp. Quant. Biol. 63, 105–110 (1998).
Schultz, P. et al. Molecular structure of human TFIIH. Cell 102, 599–607 (2000).
Lehmann, A.R. DNA repair-deficient diseases, xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. Biochimie 85, 1101–1111 (2003).
Fischer, L. et al. Cloning of the 62-kilodalton component of basic transcription factor BTF2. Science 257, 1392–1395 (1992).
Tirode, F., Busso, D., Coin, F. & Egly, J.M. Reconstitution of the transcription factor TFIIH: assignment of functions for the three enzymatic subunits, XPB, XPD, and cdk7. Mol. Cell 3, 87–95 (1999).
Ohkuma, Y. Multiple functions of general transcription factors TFIIE and TFIIH in transcription: possible points of regulation by trans-acting factors. J. Biochem. 122, 481–489 (1997).
Svejstrup, J.Q., Vichi, P. & Egly, J.M. The multiple roles of transcription/repair factor TFIIH. Trends Biochem. Sci. 21, 346–350 (1996).
Bushnell, D.A., Bamdad, C. & Kornberg, R.D. A minimal set of RNA polymerase II transcription protein interactions. J. Biol. Chem. 271, 20170–20174 (1996).
Lu, H., Zawel, L., Fisher, L., Egly, J.M. & Reinberg, D. Human general transcription factor IIH phosphorylates the C-terminal domain of RNA polymerase II. Nature 358, 641–645 (1992).
Zurita, M. & Merino, C. The transcriptional complexity of the TFIIH complex. Trends Genet. 19, 578–584 (2003).
Xiao, H. et al. Binding of basal transcription factor TFIIH to the acidic activation domains of VP16 and p53. Mol. Cell. Biol. 14, 7013–7024 (1994).
Vandel, L. & Kouzarides, T. Residues phosphorylated by TFIIH are required for E2F-1 degradation during S-phase. EMBO J. 18, 4280–4291 (1999).
Chen, D. et al. Activation of estrogen receptor alpha by S118 phosphorylation involves a ligand-dependent interaction with TFIIH and participation of CDK7. Mol. Cell 6, 127–137 (2000).
Yokoi, M. et al. The xeroderma pigmentosum group C protein complex XPC-HR23B plays an important role in the recruitment of transcription factor IIH to damaged DNA. J. Biol. Chem. 275, 9870–9875 (2000).
Iyer, N., Reagan, M.S., Wu, K.J., Canagarajah, B. & Friedberg, E.C. Interactions involving the human RNA polymerase II transcription/nucleotide excision repair complex TFIIH, the nucleotide excision repair protein XPG, and Cockayne syndrome group B (CSB) protein. Biochemistry 35, 2157–2167 (1996).
Gervais, V., Lamour, V., Gaudin, F., Thierry, J.C. & Kieffer, B. Assignment of the 1H, 15N, 13C resonances of the N-terminal domain of the human TFIIH p62 subunit. J. Biomol. NMR 19, 281–282 (2001).
Ruckert, M. & Otting, G.J. Alignment of biological macromolecules in novel nonionic liquid crystalline media for NMR experiments. J. Am. Chem. Soc. 122, 7793–7797 (2000).
Tjandra, N., Wingfield, P., Stahl, S. & Bax, A. Anisotropic rotational diffusion of perdeuterated HIV protease from 15N NMR relaxation measurements at two magnetic fields. J. Biomol. NMR 8, 273–284 (1996).
Holm, L. & Sander, C. Protein structure comparison by alignment of distance matrices. J Mol. Biol. 233, 123–138 (1993).
Blomberg, N., Baraldi, E., Nilges, M. & Saraste, M. The PH superfold: a structural scaffold for multiple functions. Trends Biochem. Sci. 24, 441–445 (1999).
Dhe-Paganon, S., Ottinger, E.A., Nolte, R.T., Eck, M.J. & Shoelson, S.E. Crystal structure of the pleckstrin homology-phosphotyrosine binding (PH-PTB) targeting region of insulin receptor substrate 1. Proc. Natl. Acad. Sci. USA 96, 8378–8383 (1999).
Pearson, M.A., Reczek, D., Bretscher, A. & Karplus, P.A. Structure of the ERM protein moesin reveals the FERM domain fold masked by an extended actin binding tail domain. Cell 101, 259–270 (2000).
Ferguson, K.M. et al. Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains. Mol. Cell 6, 373–384 (2000).
Araujo, S.J. et al. Nucleotide excision repair of DNA with recombinant human proteins: definition of the minimal set of factors, active forms of TFIIH, and modulation by CAK. Genes Dev. 14, 349–359 (2000).
Jawhari, A. et al. p52 mediates XPB function within the transcription/repair factor TFIIH. J. Biol. Chem. 277, 31761–31767 (2002).
Coin, F. et al. Mutations in the XPD helicase gene result in XP and TTD phenotypes, preventing interaction between XPD and the p44 subunit of TFIIH. Nat. Genet. 20, 184–188 (1998).
Seroz, T., Perez, C., Bergmann, E., Bradsher, J. & Egly, J.M. p44/SSL1, the regulatory subunit of the XPD/RAD3 helicase, plays a crucial role in the transcriptional activity of TFIIH. J. Biol. Chem. 275, 33260–33266 (2000).
Yan, K.S., Kuti, M. & Zhou, M.M. PTB or not PTB? that is the question. FEBS Lett. 513, 67–70 (2002).
Schmidt, A. & Hall, A. The Rho exchange factor Net1 is regulated by nuclear sequestration. J. Biol. Chem. 277, 14581–14588 (2002).
Tanaka, K. et al. Evidence that a phosphatidylinositol 3,4,5-trisphosphate-binding protein can function in nucleus. J. Biol. Chem. 274, 3919–3922 (1999).
Irvine, R.F. Nuclear lipid signalling. Nat. Rev. Mol. Cell Biol. 4, 349–360 (2003).
Cook, P.R. The organization of replication and transcription. Science 284, 1790–1795 (1999).
Araujo, S.J., Nigg, E.A. & Wood, R.D. Strong functional interactions of TFIIH with XPC and XPG in human DNA nucleotide excision repair, without a preassembled repairosome. Mol. Cell Biol. 21, 2281–2291 (2001).
Winkler, G.S., Sugasawa, K., Eker, A.P., de Laat, W.L. & Hoeijmakers, J.H. Novel functional interactions between nucleotide excision DNA repair proteins influencing the enzymatic activities of TFIIH, XPG, and ERCC1-XPF. Biochemistry 40, 160–165 (2001).
Cornilescu, G., Delaglio, F. & Bax, A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13, 289–302 (1999).
Vuister, G.W., Delaglio, F. & Bax, A. The use of 1JCα-Hα coupling constants as a probe for protein backbone conformation. J. Biomol. NMR 3, 67–80 (1993).
Ottiger, M., Delaglio, F. & Bax, A. Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra. J. Magn. Reson. 131, 373–378 (1998).
Clore, G.M., Gronenborn, A.M. & Tjandra, N. Direct structure refinement against residual dipolar couplings in the presence of rhombicity of unknown magnitude. J. Magn. Reson. 131, 159–162 (1998).
Brunger, A.T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
Nilges, M., Macias, M.J., O'Donoghue, S.I. & Oschkinat, H. Automated NOESY interpretation with ambiguous distance restraints: the refined NMR solution structure of the pleckstrin homology domain from beta-spectrin. J. Mol. Biol. 269, 408–422 (1997).
Schwieters, C.D., Kuszewski, J.J., Tjandra, N. & Clore, G.M. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 160, 65–73 (2003).
Koradi, R., Billeter, M. & Wuthrich, K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14, 51–55 (1996).
Laskowski, R.A., Rullman, J.A., MacArthur, M.W., Kaptein, R. & Thornton, J.M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8, 477–486 (1996).
Vriend, G. WHAT IF: a molecular modeling and drug design program. J. Mol. Graph. 8, 52–56 (1990).
Jawhari, A. et al. Expression of FLAG fusion proteins in insect cells: application to the multi-subunit transcription/DNA repair factor TFIIH. Protein Expr. Purif. 24, 513–523 (2002).
Gerard, M. et al. Purification and interaction properties of the human RNA polymerase B(II) general transcription factor BTF2. J. Biol. Chem. 266, 20940–20945 (1991).
Shivji, M.K., Moggs, J.G., Kuraoka, I. & Wood, R.D. Dual-incision assays for nucleotide excision repair using DNA with a lesion at a specific site. Methods Mol. Biol. 113, 373–392 (1999).
Thompson, J.D., Plewniak, F., Thierry, J.C. & Poch, O. DbClustal: rapid and reliable global multiple alignments of protein sequences detected by database searches. Nucleic Acids Res. 28, 2919–2926 (2000).
Holm, L. & Sander, C. Alignment of three-dimensional protein structures: network server for database searching. Methods Enzymol. 266, 653–662 (1996).
Clore, G.M. & Garrett, D.S. R-factor, Free R, and complete cross-validation for dipolar coupling refinement of NMR structures. J. Am. Chem. Soc. 121, 9008–9012 (1999).
Acknowledgements
This work was supported by funds from the Université Louis Pasteur de Strasbourg, the Centre National de la Recherche Scientifique, the Institut National de la Santé et de la Recherche Médicale and SPINE (contract QLG2-CT-2002-00988). V.G. was supported by a grant from ADRERUS/LILLY and by the Ministère de la Recherche et de l'Enseignement Supérieur and A.J. by the Association pour la Recherche contre le Cancer. We express our gratitude to D. Moras for constant support and fruitful discussions. We wish to acknowledge A. Milon at the IPBS in Toulouse for his hospitality during structure calculations. I. Kolb and J.L. Weickert are acknowledged for insect cell production. We are grateful to C. Ling for the management of the computing and NMR facilities and to R.A. Atkinson for critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Secondary structure elements of p62(1–108). (PDF 34 kb)
Rights and permissions
About this article
Cite this article
Gervais, V., Lamour, V., Jawhari, A. et al. TFIIH contains a PH domain involved in DNA nucleotide excision repair. Nat Struct Mol Biol 11, 616–622 (2004). https://doi.org/10.1038/nsmb782
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb782