Abstract
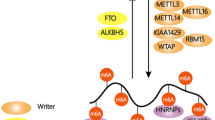
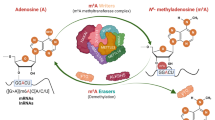
RNA modifications are integral to the regulation of RNA metabolism. One abundant mRNA modification is N6-methyladenosine (m6A), which affects various aspects of RNA metabolism, including splicing, translation and degradation. Current knowledge about the proteins recruited to m6A to carry out these molecular processes is still limited. Here we describe comprehensive and systematic mass-spectrometry-based screening of m6A interactors in various cell types and sequence contexts. Among the main findings, we identified G3BP1 as a protein that is repelled by m6A and positively regulates mRNA stability in an m6A-regulated manner. Furthermore, we identified FMR1 as a sequence-context-dependent m6A reader, thus revealing a connection between an mRNA modification and an autism spectrum disorder. Collectively, our data represent a rich resource and shed further light on the complex interplay among m6A, m6A interactors and mRNA homeostasis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Dimock, K. & Stoltzfus, C.M. Sequence specificity of internal methylation in B77 avian sarcoma virus RNA subunits. Biochemistry 16, 471–478 (1977).
Desrosiers, R.C., Friderici, K.H. & Rottman, F.M. Characterization of Novikoff hepatoma mRNA methylation and heterogeneity in the methylated 5′ terminus. Biochemistry 14, 4367–4374 (1975).
Zheng, G. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29 (2013).
Jia, G. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887 (2011).
Liu, J. et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95 (2014).
Ping, X.L. et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24, 177–189 (2014).
Schwartz, S. et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 8, 284–296 (2014).
Aguilo, F. et al. Coordination of m6A mRNA methylation and gene transcription by ZFP217 regulates pluripotency and reprogramming. Cell Stem Cell 17, 689–704 (2015).
Dominissini, D. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012).
Meyer, K.D. et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646 (2012).
Zhao, X. et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 24, 1403–1419 (2014).
Zhong, S. et al. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 20, 1278–1288 (2008).
Agarwala, S.D., Blitzblau, H.G., Hochwagen, A. & Fink, G.R. RNA methylation by the MIS complex regulates a cell fate decision in yeast. PLoS Genet. 8, e1002732 (2012).
Lin, S., Choe, J., Du, P., Triboulet, R. & Gregory, R.I. The m6A methyltransferase METTL3 promotes translation in human cancer cells. Mol. Cell 62, 335–345 (2016).
Li, Z. et al. FTO plays an oncogenic role in acute myeloid leukemia as a N6-methyladenosine RNA demethylase. Cancer Cell 31, 127–141 (2017).
Batista, P.J. et al. m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 15, 707–719 (2014).
Zhao, B.S. et al. m6A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature 542, 475–478 (2017).
Geula, S. et al. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 347, 1002–1006 (2015).
Wang, X. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120 (2014).
Meyer, K.D. et al. 5′ UTR m6A promotes cap-independent translation. Cell 163, 999–1010 (2015).
Zhou, J. et al. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 526, 591–594 (2015).
Wang, X. et al. N6-methyladenosine modulates messenger RNA translation efficiency. Cell 161, 1388–1399 (2015).
Alarcón, C.R., Lee, H., Goodarzi, H., Halberg, N. & Tavazoie, S.F. N6-methyladenosine marks primary microRNAs for processing. Nature 519, 482–485 (2015).
Xiao, W. et al. Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol. Cell 61, 507–519 (2016).
Patil, D.P. et al. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537, 369–373 (2016).
Cao, G., Li, H.B., Yin, Z. & Flavell, R.A. Recent advances in dynamic m6A RNA modification. Open Biol. 6, 160003 (2016).
Shi, H. et al. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 27, 315–328 (2017).
Alarcón, C.R. et al. HNRNPA2B1 is a mediator of m6A-dependent nuclear RNA processing events. Cell 162, 1299–1308 (2015).
Liu, N. et al. N6-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518, 560–564 (2015).
Liu, N. et al. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 45, 6051–6063 (2017).
Spruijt, C.G. & Vermeulen, M. DNA methylation: old dog, new tricks? Nat. Struct. Mol. Biol. 21, 949–954 (2014).
Penagarikano, O., Mulle, J.G. & Warren, S.T. The pathophysiology of fragile X syndrome. Annu. Rev. Genomics Hum. Genet. 8, 109–129 (2007).
Matsuki, H. et al. Both G3BP1 and G3BP2 contribute to stress granule formation. Genes Cells 18, 135–146 (2013).
Tourrière, H. et al. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 160, 823–831 (2003).
Kedersha, N. et al. G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J. Cell Biol. 212, 845–860 (2016).
Pendleton, K.E. et al. The U6 snRNA m6A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell 169, 824–835 (2017).
Castello, A. et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393–1406 (2012).
Baltz, A.G. et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell 46, 674–690 (2012).
Kwon, S.C. et al. The RNA-binding protein repertoire of embryonic stem cells. Nat. Struct. Mol. Biol. 20, 1122–1130 (2013).
Irvine, K., Stirling, R., Hume, D. & Kennedy, D. Rasputin, more promiscuous than ever: a review of G3BP. Int. J. Dev. Biol. 48, 1065–1077 (2004).
Wei, C.M. & Moss, B. Nucleotide sequences at the N6-methyladenosine sites of HeLa cell messenger ribonucleic acid. Biochemistry 16, 1672–1676 (1977).
Ke, S. et al. A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes Dev. 29, 2037–2053 (2015).
Linder, B. et al. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 12, 767–772 (2015).
Tani, H. et al. Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. Genome Res. 22, 947–956 (2012).
Myrick, L.K., Hashimoto, H., Cheng, X. & Warren, S.T. Human FMRP contains an integral tandem Agenet (Tudor) and KH motif in the amino terminal domain. Hum. Mol. Genet. 24, 1733–1740 (2015).
Brown, V. et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell 107, 477–487 (2001).
Darnell, J.C. et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146, 247–261 (2011).
Ascano, M. Jr. et al. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature 492, 382–386 (2012).
Schwanhäusser, B., Gossen, M., Dittmar, G. & Selbach, M. Global analysis of cellular protein translation by pulsed SILAC. Proteomics 9, 205–209 (2009).
Visscher, M. et al. Proteome-wide changes in protein turnover rates in C. elegans models of longevity and age-related disease. Cell Rep. 16, 3041–3051 (2016).
Chen, C.A. & Shyu, A.B. Emerging themes in regulation of global mRNA turnover in cis. Trends Biochem. Sci. 42, 16–27 (2017).
Costa, M., Ochem, A., Staub, A. & Falaschi, A. Human DNA helicase VIII: a DNA and RNA helicase corresponding to the G3BP protein, an element of the ras transduction pathway. Nucleic Acids Res. 27, 817–821 (1999).
Anderson, P. & Kedersha, N. Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 33, 141–150 (2008).
Scadden, A.D. Inosine-containing dsRNA binds a stress-granule-like complex and downregulates gene expression in trans. Mol. Cell 28, 491–500 (2007).
Kim, S.H., Dong, W.K., Weiler, I.J. & Greenough, W.T. Fragile X mental retardation protein shifts between polyribosomes and stress granules after neuronal injury by arsenite stress or in vivo hippocampal electrode insertion. J. Neurosci. 26, 2413–2418 (2006).
Spruijt, C.G. et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell 152, 1146–1159 (2013).
Qian, K. et al. A simple and efficient system for regulating gene expression in human pluripotent stem cells and derivatives. Stem Cells 32, 1230–1238 (2014).
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013).
Boersema, P.J., Raijmakers, R., Lemeer, S., Mohammed, S. & Heck, A.J. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 4, 484–494 (2009).
Pearson, W.R., Wood, T., Zhang, Z. & Miller, W. Comparison of DNA sequences with protein sequences. Genomics 46, 24–36 (1997).
Trapnell, C., Pachter, L. & Salzberg, S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009).
Corcoran, D.L. et al. PARalyzer: definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome Biol. 12, R79 (2011).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Hinrichs, A.S. et al. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 34, D590–D598 (2006).
Huang, W., Sherman, B.T. & Lempicki, R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009).
Supek, F., Bošnjak, M., Škunca, N. & Šmuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6, e21800 (2011).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Bray, N.L., Pimentel, H., Melsted, P. & Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008).
Wiśniewski, J.R., Zougman, A. & Mann, M. Combination of FASP and StageTip-based fractionation allows in-depth analysis of the hippocampal membrane proteome. J. Proteome Res. 8, 5674–5678 (2009).
Acknowledgements
We thank members of the Vermeulen lab for fruitful discussions. Work in the Vermeulen lab is supported by the NWO Gravitation program Cancer Genomics Netherlands. Work in the He lab is supported by NHGRI, NIH (HG008688). C.H. is an Investigator of the Howard Hughes Medical Institute. Work in the Carell lab is supported by Deutsche Forschungsgemeinschaft (grants SFB749, SFB1032 and SPP1784) and Bundesministerium für Bildung und Forschung (EXC114).
Author information
Authors and Affiliations
Contributions
R.R.E. and M.V. conceived the project and wrote the manuscript, with input from all other authors. R.R.E. performed most wet-lab experiments. S.G. and M.R. prepared the m6A probes. R.G.H.L. analyzed RNA-seq and whole cell proteome data. H.S. and P.J.H. performed CLIP and PAR-CLIP experiments. Z.L. analyzed PAR-CLIP data. S.-Y.W. performed bioinformatics analysis of FMR1 PAR-CLIP data. M.P.A.B. provided technical support. P.W.T.C.J. measured mass spectrometry samples. M.M. and H.G.S. provided scientific input. C.H., T.C. and M.V. supervised the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 A global m6A interactome.
(a) Schematic representation of the workflow used in this study (see detailed description of the approach and analysis in Online Methods). (b) Structure of the biotin molecule and linker used in the m6A probes. The RNA strand is attached to the phosphate group that is shown. (c) Mass spectra showing control and m6A probes. Calculated and observed molecular weights of the probes are indicated. The observed single peaks illustrate the purity of the probes. (d) Agarose gel showing depletion of biotin tagged probes after incubation with streptavidin beads. (e-f) Scatter plots of SILAC-based m6A pull-downs in mouse ESC nuclear extract I and mouse ESC cytoplasmic extract (f). (g-h) Scatter plots of di-methyl based m6A pull-downs in mNPC nuclear extract (g) and mNPC cytoplasmic extract (h). (i) Scatter plots of m6A pull-downs in mouse 3T3 nuclear lysates. (j-k) Scatter plots of SILAC based m6A pull downs in mESC whole cell extract using degenerate sequence probe 1 (j) and degenerate sequence probe 2 (k) The sequence context of the probes is depicted at the bottom. (l) Protein domain distribution of m6A repelled proteins in humans. (related to Fig. 1).
Supplementary Figure 2 G3BP1 and G3BP2 PAR-CLIP analysis.
(a) Scatter plot of a SILAC-based m6A pull down in mESC cytoplasmic extract in a GAm6ACU sequence context. Note the presence of G3BP1 in the background cloud. (b) The summary of PAR-CLIP sequencing reads (c) The genomic distribution of G3BP1 and G3BP2 binding sites on mRNA. (d) Enriched Most GO-terms for G3BP2 target genes. (e-f) Distribution of G3BP1 (e) and G3BP2 (f) peaks relative to YTHDF2. (g-h) Distribution of G3BP1 (g) and G3BP2 (h) peaks relative to YTHDC1 across the transcriptome. (related to Fig. 2).
Supplementary Figure 3 RNA-seq validation.
(a) Correlation between YTHDF2 mRNA binding and mRNA stability after the effect of m6A on mRNA stability was regressed out (see Online Methods). (b) Q-PCR validation showing the effect of G3BP1 KD on mRNA stability (Data are shown as range; n = 2 independent biological experiments). (c) Q-PCR validation showing the effect of G3BP1 OE on mRNA stability (Data are shown as means ± data range; n = 2 independent experiments) (related to Fig. 3).
Supplementary Figure 4 In vitro and in vivo data regarding FMR1 and m6A.
(a) FMR1 domain structure and overview of different GST-tagged FMR1 deletion constructs (b) GST western blot analysis of GST-FMR1 deletion constructs binding to unmodified and m6A-containing RNA probes (GGACU context). For all constructs, the correct size band is indicated with a red arrow. (c) Western blots showing expression of FLAG-HA, FLAG-HA-FMR1 and FLAG-HA-FMR1 I304N mutant (FMR1-m) in HEK293 cells. (d) Representative LC/MS based quantification showing levels of m6A in FLAG-HA-FMR1 and FLAG-HA-FMR1 mutant enriched fraction, prior to RiboMinus treatment. Data are shown as range; n = 2 independent biological experiments. (e) Venn diagram showing the overlap between FMR1 (mouse brain) and YTHDF1 (HEK cells) target transcripts. (f) GO-term enrichment analysis of common target transcripts of FMR1 (mouse brain) and YTHDF1 (HEK cells). GO terms related to neurogenesis are indicated in red. (g) Box-plots depicting the global changes in protein half-lives in the indicated samples. (related to Fig. 4).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 (PDF 1022 kb)
Supplementary Table 1
RNA probes and oligos (XLSX 12 kb)
Supplementary Data Set 1
Methods and uncropped images. (PDF 8355 kb)
Supplementary Data Set 2
Mass spectrometry data from RNA pulldowns. (XLSX 6909 kb)
Supplementary Data Set 3
G3BP1 and G3BP2 PAR-CLIP data. (XLSX 3279 kb)
Supplementary Data Set 4
mRNA half-life estimation RNA-seq data. (XLSX 623 kb)
Supplementary Data Set 5
Pulse-SILAC data. (XLSX 725 kb)
Rights and permissions
About this article
Cite this article
Edupuganti, R., Geiger, S., Lindeboom, R. et al. N6-methyladenosine (m6A) recruits and repels proteins to regulate mRNA homeostasis. Nat Struct Mol Biol 24, 870–878 (2017). https://doi.org/10.1038/nsmb.3462
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.3462
This article is cited by
-
Transcriptome-wide dynamics of m6A methylation in ISKNV and Siniperca chuatsi cells infected with ISKNV
BMC Genomics (2025)
-
Developing novel Lin28 inhibitors by computer aided drug design
Cell Death Discovery (2025)
-
Aberrant splicing in Huntington’s disease accompanies disrupted TDP-43 activity and altered m6A RNA modification
Nature Neuroscience (2025)
-
Recruitment of the m6A/m6Am demethylase FTO to target RNAs by the telomeric zinc finger protein ZBTB48
Genome Biology (2024)
-
mRNA m6A detection
Nature Reviews Methods Primers (2024)