Abstract
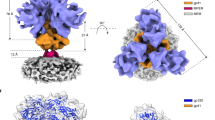
The activation of trimeric HIV-1 envelope glycoprotein (Env) by its binding to the cell-surface receptor CD4 and co-receptors (CCR5 or CXCR4) represents the first of a series of events that lead to fusion between viral and target-cell membranes. Here, we present the cryo-EM structure, at subnanometer resolution (∼6 Å at 0.143 FSC), of the 'closed', prefusion state of trimeric HIV-1 Env complexed to the broadly neutralizing antibody VRC03. We show that three gp41 helices at the core of the trimer serve as an anchor around which the rest of Env is reorganized upon activation to the 'open' quaternary conformation. The architecture of trimeric HIV-1 Env in the prefusion state and in the activated intermediate state resembles the corresponding states of influenza hemagglutinin trimers, thus providing direct evidence for the similarity in entry mechanisms used by HIV-1, influenza and related enveloped viruses.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
10 November 2013
In the version of this article initially published online, the NIH-FEI Living Lab for Structural Biology was not mentioned in the Acknowledgments section. The error has been corrected for the print, PDF and HTML versions of this article.
References
Wyatt, R. & Sodroski, J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280, 1884–1888 (1998).
Kwong, P.D. et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393, 648–659 (1998).
Huang, C.C. et al. Structure of a V3-containing HIV-1 gp120 core. Science 310, 1025–1028 (2005).
Zhou, T. et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445, 732–737 (2007).
Pancera, M. et al. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc. Natl. Acad. Sci. USA 107, 1166–1171 (2010).
Zhou, T. et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329, 811–817 (2010).
Merk, A. & Subramaniam, S. HIV-1 envelope glycoprotein structure. Curr. Opin. Struct. Biol. 23, 268–276 (2013).
Chan, D.C., Fass, D., Berger, J.M. & Kim, P.S. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89, 263–273 (1997).
Weissenhorn, W., Dessen, A., Harrison, S.C., Skehel, J.J. & Wiley, D.C. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387, 426–430 (1997).
Bartesaghi, A. et al. Classification and 3D averaging with missing wedge correction in biological electron tomography. J. Struct. Biol. 162, 436–450 (2008).
Frank, G.A. et al. Computational separation of conformational heterogeneity using cryo-electron tomography and 3D sub-volume averaging. J. Struct. Biol. 178, 165–176 (2012).
Liu, J., Bartesaghi, A., Borgnia, M.J., Sapiro, G. & Subramaniam, S. Molecular architecture of native HIV-1 gp120 trimers. Nature 455, 109–113 (2008).
Meyerson, J.R. et al. Molecular structures of trimeric HIV-1 Env in complex with small antibody derivatives. Proc. Natl. Acad. Sci. USA 110, 513–518 (2013).
Tran, E.E. et al. Structural mechanism of trimeric HIV-1 envelope glycoprotein activation. PLoS Pathog. 8, e1002797 (2012).
Harris, A. et al. Trimeric HIV-1 glycoprotein gp140 immunogens and native HIV-1 envelope glycoproteins display the same closed and open quaternary molecular architectures. Proc. Natl. Acad. Sci. USA 108, 11440–11445 (2011).
Sanders, R.W. et al. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J. Virol. 76, 8875–8889 (2002).
Moscoso, C.G. et al. Quaternary structures of HIV Env immunogen exhibit conformational vicissitudes and interface diminution elicited by ligand binding. Proc. Natl. Acad. Sci. USA 108, 6091–6096 (2011).
Wu, S.R. et al. Single-particle cryoelectron microscopy analysis reveals the HIV-1 spike as a tripod structure. Proc. Natl. Acad. Sci. USA 107, 18844–18849 (2010).
Wu, S. et al. Fabs enable single particle cryoEM studies of small proteins. Structure 20, 582–592 (2012).
Grigorieff, N. FREALIGN: high-resolution refinement of single particle structures. J. Struct. Biol. 157, 117–125 (2007).
Scheres, S.H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Pettersen, E.F. et al. UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Wu, X. et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 333, 1593–1602 (2011).
Mao, Y. et al. Molecular architecture of the uncleaved HIV-1 envelope glycoprotein trimer. Proc. Natl. Acad. Sci. USA 110, 12438–12443 (2013).
Kwon, Y.D. et al. Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops. Proc. Natl. Acad. Sci. USA 109, 5663–5668 (2012).
Fontana, J., Cardone, G., Heymann, J.B., Winkler, D.C. & Steven, A.C. Structural changes in Influenza virus at low pH characterized by cryo-electron tomography. J. Virol. 86, 2919–2929 (2012).
Xu, R. & Wilson, I.A. Structural characterization of an early fusion intermediate of influenza virus hemagglutinin. J. Virol. 85, 5172–5182 (2011).
Harrison, S.C. Viral membrane fusion. Nat. Struct. Mol. Biol. 15, 690–698 (2008).
Weissenhorn, W., Carfi, A., Lee, K.H., Skehel, J.J. & Wiley, D.C. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol. Cell 2, 605–616 (1998).
Wilson, I.A., Skehel, J.J. & Wiley, D.C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature 289, 366–373 (1981).
Bullough, P.A., Hughson, F.M., Skehel, J.J. & Wiley, D.C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371, 37–43 (1994).
Ivanovic, T., Choi, J.L., Whelan, S.P., van Oijen, A.M. & Harrison, S.C. Influenza-virus membrane fusion by cooperative fold-back of stochastically induced hemagglutinin intermediates. eLife 2, e00333 (2013).
Eckert, D.M. & Kim, P.S. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70, 777–810 (2001).
Huang, J. et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 491, 406–412 (2012).
Corti, D. & Lanzavecchia, A. Broadly neutralizing antiviral antibodies. Annu. Rev. Immunol. 31, 705–742 (2013).
McLellan, J.S. et al. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science 340, 1113–1117 (2013).
Baquero, E. et al. Intermediate conformations during viral fusion glycoprotein structural transition. Curr. Opin. Virol. 3, 143–150 (2013).
Li, X. et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods 10, 584–590 (2013).
Weis, W.I., Brunger, A.T., Skehel, J.J. & Wiley, D.C. Refinement of the influenza virus hemagglutinin by simulated annealing. J. Mol. Biol. 212, 737–761 (1990).
Kremer, J.R., Mastronarde, D.N. & McIntosh, J.R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 (1996).
Mindell, J.A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003).
Ludtke, S.J., Baldwin, P.R. & Chiu, W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 (1999).
van Heel, M. et al. in International Tables for Crystallography Volume F: Crystallography of Biological Macromolecules 2nd edn (eds. Arnold, E., Himmel, D.M. & Rossmann, M.G.) 624–628 (Wiley, 2012).
Henderson, R. et al. Tilt-pair analysis of images from a range of different specimens in single-particle electron cryomicroscopy. J. Mol. Biol. 413, 1028–1046 (2011).
Rosenthal, P.B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003).
Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007).
Acknowledgements
This work was supported by funds to S.S. and J.L.S.M. from the Center for Cancer Research at the National Cancer Institute, US National Institutes of Health (NIH), and to S.S. from the NIH Intramural AIDS Targeted Antiviral Program. We thank J. Mascola (Vaccine Research Center, NIH) for providing VRC03 antibodies; K. Kang and W. Olson (Progenics) for providing soluble KNH1144 gp140 trimers; S. Fellini, S. Chacko and their colleagues for continued support with use of the Biowulf cluster for computing at NIH; D. Schauder and H. He for assistance with data collection; P. Rao and the NIH-FEI Living Lab for Structural Biology for assistance with collection of the tilt-pair images; and L. Earl for helpful discussions and comments.
Author information
Authors and Affiliations
Contributions
A.B., A.M., M.J.B., J.L.S.M. and S.S. analyzed and interpreted data; S.S. was responsible for data collection; all authors helped compose the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
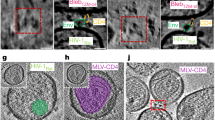
Supplementary Figure 1 Cryo-electron microscopic imaging of the Env-VRC03 complex.
(a, b) Representative images recorded at a dose of ∼10 electrons/Å2 and underfocus values of ∼2.6 m (a) and ∼1.5 m (b) (leftmost panels). Scale bar is 400 Å Band-pass filtered versions of these images allows easier particle visualization (middle panels). Rotationally averaged FFTs of each image show signal from the Thon rings extending to ∼8-Å for micrographs at higher defocus values and to ∼6-Å for micrographs at lower defocus values (rightmost panels). (c) Gallery of selected initial class averages obtained by classification of single particle projection images without any prior translational or rotational alignment reveals the characteristic views of the VRC03-bound gp120 trimer.
Supplementary Figure 2 Map resolution and validation.
(a, b) Maps obtained from FREALIGN20 (a) and RELION21 (b) shown in top view, fitted with PDB 3SE8 (ref. 23) coordinates. (c) Plots of the Fourier Shell Correlation (FSC) coefficient for the structure of the Env-VRC03 complex. The curves show the “gold-standard” curve obtained using RELION and the curve obtained from the correlation of two halves of the data set obtained using FREALIGN, both indicating a resolution value of ∼6-Å as measured by the 0.143 FSC cutoff criterion. (d) Validation of density map using tilt-pair parameter plot as suggested by R. Henderson and colleagues44. The spread in orientational assignments around the known goniometer settings is within ∼12.5° for >62% of the selected particle pairs, consistent with the ∼6-Å resolution reported for the maps.
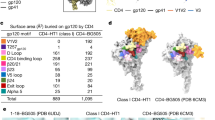
Supplementary Figure 3 Correspondence between 6-Å and 20-Å density maps.
(a) 20-Å structure of the complex of VRC03 with native trimeric Env obtained using cryo-electron tomography of intact HIV-1 (ref. 14). Side views (left) and top views (right) of the density map, fitted with three copies of the structure of the complex of VRC03 Fab with gp120 (PDB 3SE8 (ref. 23)). The 2D cryo-electron microscopic images in Figure 1 can be recognized as projections of the propeller-shaped 3D structure of the Env-VRC03 complex shown here. (b) Side (left) and top views (right) of the superposition of the density map of soluble trimeric HIV-1 Env-VRC03 Fab complex (obtained using cryo-electron microscopy at ∼6-Å resolution) with density map of the native trimeric HIV-1 Env-VRC03 complex (obtained using cryo-electron tomography at ∼20-Å resolution).
Supplementary Figure 4 Change in appearance of density map with filtering to lower resolutions.
(a-c) Side views of the 6-Å density of map of the complex of VRC03 Fab with soluble trimeric HIV-1 Env, filtered to resolutions of 10-Å (a), 15-Å (b) and 20-Å (c). As the resolution is progressively lowered, the overall shape of the map is unaltered, but the central helices which are prominently resolved at 10-Å resolution, fade into the background at resolutions of 20-Å or lower, resulting in what has the appearance of a central cavity that is observed in the tomographic density maps of unliganded and VRC03-bound trimeric Env.
Supplementary Figure 6 Structure of the open, activated trimeric Env conformation.
Side (top) and top view (bottom) of the 9-Å density map of the open state of trimeric Env stabilized in a complex with the Fab fragment of the 17b monoclonal antibody. The map is fitted with three copies of PDB 3HMG (ref. 39) for the gp41 central densities (cyan) and three copies of gp120 (red) and the Fv fragment of 17b (green) derived from the coordinates for the structure of the gp120-sCD4-17b complex (PDB 1GC1 (ref. 2)).
Supplementary information
Supplementary Figures
Supplementary Figures 1–6 (PDF 7787 kb)
Rights and permissions
About this article
Cite this article
Bartesaghi, A., Merk, A., Borgnia, M. et al. Prefusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy. Nat Struct Mol Biol 20, 1352–1357 (2013). https://doi.org/10.1038/nsmb.2711
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2711