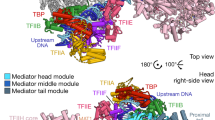
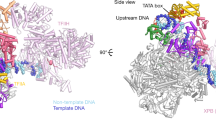
Abstract
We used single-particle electron microscopy to characterize the structure and subunit organization of the Mediator Head module that controls Mediator–RNA polymerase II (RNAPII) and Mediator-promoter interactions. The Head module adopts several conformations differing in the position of a movable jaw formed by the Med18–Med20 subcomplex. We also characterized, by structural, biochemical and genetic means, the interactions of the Head module with TATA-binding protein (TBP) and RNAPII subunits Rpb4 and Rpb7. TBP binds near the Med18–Med20 attachment point and stabilizes an open conformation of the Head module. Rpb4 and Rpb7 bind between the Head jaws, establishing contacts essential for yeast-cell viability. These results, and consideration of the structure of the Mediator–RNAPII holoenzyme, shed light on the stabilization of the pre-initiation complex by Mediator and suggest how Mediator might influence initiation by modulating polymerase conformation and interaction with promoter DNA.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Flanagan, P.M. et al. Resolution of factors required for the initiation of transcription by yeast RNA polymerase II. J. Biol. Chem. 265, 11105–11107 (1990).
Flanagan, P.M., Kelleher, R.J. III., Sayre, M.H., Tschochner, H. & Kornberg, R.D. A mediator required for activation of RNA polymerase II transcription in vitro. Nature 350, 436–438 (1991).
Baek, H.J., Malik, S., Qin, J. & Roeder, R.G. Requirement of TRAP/mediator for both activator-independent and activator-dependent transcription in conjunction with TFIID-associated TAF(II)s. Mol. Cell. Biol. 22, 2842–2852 (2002).
Naar, A.M. et al. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature 398, 828–832 (1999).
Takagi, Y. & Kornberg, R.D. Mediator as a general transcription factor. J. Biol. Chem. 281, 80–89 (2006).
Asturias, F.J., Jiang, Y.W., Myers, L.C., Gustafsson, C.M. & Kornberg, R.D. Conserved structures of mediator and RNA polymerase II holoenzyme. Science 283, 985–987 (1999).
Sato, S. et al. A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol. Cell 14, 685–691 (2004).
Cai, G., Imasaki, T., Takagi, Y. & Asturias, F.J. Mediator structural conservation and implications for the regulation mechanism. Structure 17, 559–567 (2009).
Holstege, F.C. et al. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95, 717–728 (1998).
Thompson, C.M. & Young, R.A. General requirement for RNA polymerase II holoenzymes in vivo. Proc. Natl. Acad. Sci. USA 92, 4587–4590 (1995).
Takagi, Y. et al. Head module control of mediator interactions. Mol. Cell 23, 355–364 (2006).
Fitzgerald, D.J. et al. Protein complex expression by using multigene baculoviral vectors. Nat. Methods 3, 1021–1032 (2006).
Radermacher, M. The three-dimensional reconstruction of single particles from random and non-random tilt series. J. Electron Microsc. Tech. 9, 359–394 (1988).
Frank, J. Three-Dimensional Electron Microscopy of Macromolecular Assemblies 410 (Oxford University Press, San Diego, California, USA, 2006).
Lariviere, L. et al. Structure and TBP binding of the Mediator head subcomplex Med8-Med18-Med20. Nat. Struct. Mol. Biol. 13, 895–901 (2006).
Koh, S.S., Ansari, A.Z., Ptashne, M. & Young, R.A. An activator target in the RNA polymerase II holoenzyme. Mol. Cell 1, 895–904 (1998).
Toth-Petroczy, A. et al. Malleable machines in transcription regulation: the mediator complex. PLOS Comput. Biol. 4, e1000243 (2008).
Baumli, S., Hoeppner, S. & Cramer, P. A conserved mediator hinge revealed in the structure of the MED7.MED21 (Med7.Srb7) heterodimer. J. Biol. Chem. 280, 18171–18178 (2005).
Koschubs, T. et al. Identification, structure, and functional requirement of the Mediator submodule Med7N/31. EMBO J. 28, 69–80 (2009).
Taatjes, D.J., Schneider-Poetsch, T. & Tjian, R. Distinct conformational states of nuclear receptor-bound CRSP-Med complexes. Nat. Struct. Mol. Biol. 11, 664–671 (2004).
Bushnell, D.A. & Kornberg, R.D. Complete, 12-subunit RNA polymerase II at 4.1-Å resolution: implications for the initiation of transcription. Proc. Natl. Acad. Sci. USA 100, 6969–6973 (2003).
Armache, K.J., Kettenberger, H. & Cramer, P. Architecture of initiation-competent 12-subunit RNA polymerase II. Proc. Natl. Acad. Sci. USA 100, 6964–6968 (2003).
Davis, J.A., Takagi, Y., Kornberg, R.D. & Asturias, F.A. Structure of the yeast RNA polymerase II holoenzyme: Mediator conformation and polymerase interaction. Mol. Cell 10, 409–415 (2002).
Bushnell, D.A., Westover, K.D., Davis, R.E. & Kornberg, R.D. Structural basis of transcription: an RNA polymerase II-TFIIB cocrystal at 4.5 Angstroms. Science 303, 983–988 (2004).
Chung, W.H. et al. RNA polymerase II/TFIIF structure and conserved organization of the initiation complex. Mol. Cell 12, 1003–1013 (2003).
Chen, H.T. & Hahn, S. Mapping the location of TFIIB within the RNA polymerase II transcription preinitiation complex: a model for the structure of the PIC. Cell 119, 169–180 (2004).
Todone, F., Brick, P., Werner, F., Weinzierl, R.O. & Onesti, S. Structure of an archaeal homolog of the eukaryotic RNA polymerase II RPB4/RPB7 complex. Mol. Cell 8, 1137–1143 (2001).
Sampath, V. & Sadhale, P. Rpb4 and Rpb7: a sub-complex integral to multi-subunit RNA polymerases performs a multitude of functions. IUBMB Life 57, 93–102 (2005).
Edwards, A.M., Kane, C.M., Young, R.A. & Kornberg, R.D. Two dissociable subunits of yeast RNA polymerase II stimulate the initiation of transcription at a promoter in vitro. J. Biol. Chem. 266, 71–75 (1991).
Orlicky, S.M., Tran, P.T., Sayre, M.H. & Edwards, A.M. Dissociable Rpb4-Rpb7 subassembly of rna polymerase II binds to single-strand nucleic acid and mediates a post-recruitment step in transcription initiation. J. Biol. Chem. 276, 10097–10102 (2001).
Sampath, V., Balakrishnan, B., Verma-Gaur, J., Onesti, S. & Sadhale, P.P. Unstructured N terminus of the RNA polymerase II subunit Rpb4 contributes to the interaction of Rpb4.Rpb7 subcomplex with the core RNA polymerase II of Saccharomyces cerevisiae. J. Biol. Chem. 283, 3923–3931 (2008).
Armache, K.J., Mitterweger, S., Meinhart, A. & Cramer, P. Structures of complete RNA polymerase II and its subcomplex, Rpb4/7. J. Biol. Chem. 280, 7131–7134 (2005).
Kim, Y., Geiger, J.H., Hahn, S. & Sigler, P.B. Crystal structure of a yeast TBP/TATA-box complex. Nature 365, 512–520 (1993).
Li, M.Z. & Elledge, S.J. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat. Methods 4, 251–256 (2007).
Brignole, E.J., Smith, S. & Asturias, F.J. Conformational flexibility of metazoan fatty acid synthase enables catalysis. Nat. Struct. Mol. Biol. 16, 190–197 (2009).
Voss, N.R., Yoshioka, C.K., Radermacher, M., Potter, C.S. & Carragher, B. DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J. Struct. Biol. 166, 205–213 (2009).
Frank, J. et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 (1996).
Sorzano, C.O. et al. XMIPP: a new generation of an open-source image processing package for electron microscopy. J. Struct. Biol. 148, 194–204 (2004).
Pettersen, E.F. et al. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Sakurai, H., Mitsuzawa, H., Kimura, M. & Ishihama, A. The Rpb4 subunit of fission yeast Schizosaccharomyces pombe RNA polymerase II is essential for cell viability and similar in structure to the corresponding subunits of higher eukaryotes. Mol. Cell. Biol. 19, 7511–7518 (1999).
Acknowledgements
We thank I. Berger (European Molecular Biology Laboratory France) for providing reagents and help to establish the MultiBac expression system and C. Kaplan (Texas A&M Univ.), G. Hartzog (Univ. of California, Santa Cruz) and M. Goebl (Indiana Univ.) for providing yeast vectors and advising on the implementation of genetic experiments. This work was supported by US National Institutes of Health grant R01 GM67167 (F.J.A.) and US National Science Foundation grant MCB 0843026 (Y.T.), an American Heart Association Scientist Development Award 0735395N (Y.T.) and a Human Frontier Science Program long-term fellowship (T.I.).
Author information
Authors and Affiliations
Contributions
T.I., F.C. and Y.T. expressed, purified and biochemically characterized recombinant Head module and Head module subcomplexes and provided recombinant Rpb4 and Rpb7 and TBP; Y.T. and T.I. designed and carried out Head–Rpb4–Rpb7 and Head–TBP binding assays; K.Y. and Y.T. designed and carried out assays to test genetic interaction of Mediator subunits with Rpb4; Y.T. carried out in vitro transcription assays; G.C. carried out all EM data collection and analysis; G.C., Y.T. and F.J.A. discussed and interpreted all results; F.J.A. supervised EM structural analysis and wrote the manuscript in collaboration with G.C. and Y.T.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 and Supplementary Table 1 (PDF 352 kb)
Rights and permissions
About this article
Cite this article
Cai, G., Imasaki, T., Yamada, K. et al. Mediator Head module structure and functional interactions. Nat Struct Mol Biol 17, 273–279 (2010). https://doi.org/10.1038/nsmb.1757
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1757
This article is cited by
-
Role of Mediator in virulence and antifungal drug resistance in pathogenic fungi
Current Genetics (2019)
-
Transcription regulation by the Mediator complex
Nature Reviews Molecular Cell Biology (2018)
-
Sub1/PC4, a multifaceted factor: from transcription to genome stability
Current Genetics (2017)
-
The X-ray crystal structure of the euryarchaeal RNA polymerase in an open-clamp configuration
Nature Communications (2014)
-
Redefining the modular organization of the core Mediator complex
Cell Research (2014)