Abstract
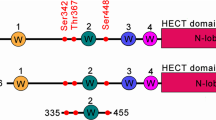
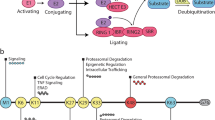
Phosphorylation and small ubiquitin-like modifier (SUMO) conjugation contribute to the spatial and temporal regulation of substrates containing phosphorylation-dependent SUMO consensus motifs (PDSMs). Myocyte-enhancement factor 2 (MEF2) is a transcription factor and PDSM substrate whose modification by SUMO drives postsynaptic dendritic differentiation. NMR analysis revealed that the human SUMO E2 interacted with model substrates for phosphorylated and nonphosphorylated MEF2 in similar extended conformations. Mutational and biochemical analysis identified a basic E2 surface that enhanced SUMO conjugation to phosphorylated PDSM substrates MEF2 and heat-shock transcription factor 1 (HSF1), but not to nonphosphorylated MEF2 or HSF1, nor the non-PDSM substrate p53. Mutant ubiquitin-conjugating enzyme UBC9 isoforms defective in promoting SUMO conjugation to phosphorylated MEF2 in vitro and in vivo also impair postsynaptic differentiation in organotypic cerebellar slices. These data support an E2-dependent mechanism that underlies phosphorylation-dependent SUMO conjugation in pathways that range from the heat-shock response to nuclear hormone signaling to brain development.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Johnson, E.S. Protein modification by SUMO. Annu. Rev. Biochem. 73, 355–382 (2004).
Geiss-Friedlander, R. & Melchior, F. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8, 947–956 (2007).
Dasso, M. Emerging roles of the SUMO pathway in mitosis. Cell Div. 3, 5 (2008).
Hershko, A. & Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 (1998).
Dye, B.T. & Schulman, B.A. Structural mechanism underlying posttranslational modification by ubiquitin-like proteins. Annu. Rev. Biophys. Biomol. Struct. 36, 131–150 (2007).
Capili, A.D. & Lima, C.D. Taking it step by step: mechanistic insights from structural studies of ubiquitin/ubiquitin-like protein modification pathways. Curr. Opin. Struct. Biol. 17, 726–735 (2007).
Kerscher, O., Felberbaum, R. & Hochstrasser, M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22, 159–180 (2006).
Sampson, D.A., Wang, M. & Matunis, M.J. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J. Biol. Chem. 276, 21664–21669 (2001).
Bernier-Villamor, V., Sampson, D.A., Matunis, M.J. & Lima, C.D. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108, 345–356 (2002).
Lin, D. et al. Identification of a substrate recognition site on Ubc9. J. Biol. Chem. 277, 21740–21748 (2002).
Reverter, D. & Lima, C.D. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature 435, 687–692 (2005).
Yunus, A.A. & Lima, C.D. Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway. Nat. Struct. Mol. Biol. 13, 491–499 (2006).
Pfander, B., Moldovan, G.L., Sacher, M., Hoege, C. & Jentsch, S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436, 428–433 (2005).
Papouli, E. et al. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol. Cell 19, 123–133 (2005).
Lin, D.Y. et al. Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol. Cell 24, 341–354 (2006).
Meulmeester, E., Kunze, M., Hsiao, H.H., Urlaub, H. & Melchior, F. Mechanism and consequences for paralog-specific SUMOylation of ubiquitin-specific protease 25. Mol. Cell 30, 610–619 (2008).
Mohideen, F. & Lima, C.D. SUMO takes control of a ubiquitin-specific protease. Mol. Cell 30, 539–540 (2008).
Gong, X. et al. Cdk5-mediated inhibition of the protective effects of transcription factor MEF2 in neurotoxicity-induced apoptosis. Neuron 38, 33–46 (2003).
Kang, J., Gocke, C.B. & Yu, H. Phosphorylation-facilitated sumoylation of MEF2C negatively regulates its transcriptional activity. BMC Biochem. 7, 5 (2006).
Yang, X.J. & Gregoire, S. A recurrent phospho-sumoyl switch in transcriptional repression and beyond. Mol. Cell 23, 779–786 (2006).
Hietakangas, V. et al. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc. Natl. Acad. Sci. USA 103, 45–50 (2006).
Ohshima, T., Koga, H. & Shimotohno, K. Transcriptional activity of peroxisome proliferator-activated receptor γ is modulated by SUMO-1 modification. J. Biol. Chem. 279, 29551–29557 (2004).
Shalizi, A. et al. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science 311, 1012–1017 (2006).
Lyons, G.E., Micales, B.K., Schwarz, J., Martin, J.F. & Olson, E.N. Expression of Mef2 genes in the mouse central nervous system suggests a role in neuronal maturation. J. Neurosci. 15, 5727–5738 (1995).
Shalizi, A. et al. PIASx is a MEF2 SUMO E3 ligase that promotes postsynaptic dendritic morphogenesis. J. Neurosci. 27, 10037–10046 (2007).
Grégoire, S. et al. Control of MEF2 transcriptional activity by coor-dinated phosphorylation and sumoylation. J. Biol. Chem. 281, 4423–4433 (2006).
Yamashita, D. et al. The transactivating function of peroxisome proliferator-activated receptor γ is negatively regulated by SUMO conjugation in the amino-terminal domain. Genes Cells 9, 1017–1029 (2004).
Yang, S.H., Galanis, A., Witty, J. & Sharrocks, A.D. An extended consensus motif enhances the specificity of substrate modification by SUMO. EMBO J. 25, 5083–5093 (2006).
Riquelme, C., Barthel, K.K.B. & Liu, X. SUMO-1 modification of MEF2A regulates its transcriptional activity. J. Cell. Mol. Med. 10, 132–144 (2006).
Verdecia, M.A., Bowman, M.E., Lu, K.P., Hunter, T. & Noel, J.P. Structural basis for phosphoserine-proline recognition by group IV WW domains. Nat. Struct. Biol. 7, 639–643 (2000).
Yunus, A.A. & Lima, C.D. Purification and activity assays for Ubc9, the ubiquitin conjugating enzyme for the small ubiquitin-like modifier SUMO. Methods Enzymol. 398, 74–87 (2005).
Lois, L.M. & Lima, C.D. Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. EMBO J. 24, 439–451 (2005).
Yunus, A.A. & Lima, C.D. Purification of SUMO conjugating enzymes and kinetic analysis of substrate conjugation. Methods Mol. Biol. 497, 167–186 (2009).
Delaglio, F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995).
Johnson, B.A. & Blevins, R.A. NMR View: a computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4, 603–614 (1994).
Liu, Q., Shen, B., Chen, D.J. & Chen, Y. Backbone resonance assignments of human UBC9. J. Biomol. NMR 13, 89–90 (1999).
Edwards, T.A. et al. Solution structure of the Vts1 SAM domain in the presence of RNA. J. Mol. Biol. 356, 1065–1072 (2006).
Gaudillière, B., Konishi, Y., de la Iglesia, N., Yao, G. & Bonni, A.A. CaMKII-NeuroD signaling pathway specifies dendritic morphogenesis. Neuron 41, 229–241 (2004).
Acknowledgements
We thank the NMR staff, particularly K. Dutta, at the New York Structural Biology Center (NYSBC) for assistance in NMR data collection and processing. We also thank A. Shalizi and A. Yunus for helpful discussions, and N. Arango for help with 293T transfection assays. NMR resources at NYSBC are supported by the US National Institutes of Health (NIH) grant P41 GM66354. The work was supported in part by NIH grants GM075695 (A.D.C.), NS041021 (A.B.) and GM065872 (C.D.L.).
Author information
Authors and Affiliations
Contributions
F.M. designed, performed and interpreted the experiments in Figures 1,2,3,4,5 and in the Supplementary Information, except for NMR experiments; A.D.C. designed, performed and interpreted NMR experiments; P.M.B. and T.Y. designed, performed and interpreted the experiments in Figure 6; A.B. supervised the research by P.M.B. and T.Y. and made contributions intellectually and to manuscript editing; C.D.L. supervised F.M. and A.D.C., designed experiments and interpreted data; F.M. and C.D.L. wrote the manuscript.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 (PDF 1265 kb)
Rights and permissions
About this article
Cite this article
Mohideen, F., Capili, A., Bilimoria, P. et al. A molecular basis for phosphorylation-dependent SUMO conjugation by the E2 UBC9. Nat Struct Mol Biol 16, 945–952 (2009). https://doi.org/10.1038/nsmb.1648
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1648