Abstract
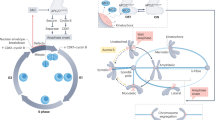
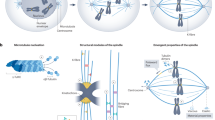
Eukaryotic cell division uses morphologically different forms of mitosis, referred to as open, partially open and closed mitosis, for accurate chromosome segregation and proper partitioning of other cellular components such as endomembranes and cell fate determinants. Recent studies suggest that the spindle matrix provides a conserved strategy to coordinate the segregation of genetic material and the partitioning of the rest of the cellular contents in all three forms of mitosis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Persico, A., Cervigni, R. I., Barretta, M. L. & Colanzi, A. Mitotic inheritance of the Golgi complex. FEBS Lett. 583, 3857–3862 (2009).
De Souza, C. P. & Osmani, S. A. Double duty for nuclear proteins — the role of more open forms of mitosis. Trends Genet. 25, 545–554 (2009).
Goldman, R. D. & Rebhun, L. I. The structure and some properties of the isolated mitotic apparatus. J. Cell Sci. 4, 179–209 (1969).
Forer, A. & Goldman, R. D. Comparisons of isolated and in vivo mitotic apparatuses. Nature 222, 689–690 (1969).
Leslie, R. J., Hird, R. B., Wilson, L., McIntosh, J. R. & Scholey, J. M. Kinesin is associated with a nonmicrotubule component of sea urchin mitotic spindle. Proc. Natl Acad. Sci. USA 84, 2771–2775 (1987).
Wein, H., Bass, H. W. & Cande, W. Z. DSK1, a kinesin-related protein involved in anaphase spindle elongation, is a component of a mitotic spindle matrix. Cell. Motil. Cytoskel. 41, 214–224 (1998).
Dumont, S. & Mitchison, T. J. Forces and length in the mitotic spindle. Curr. Biol. 19, R749–R761 (2009).
Pickett-Heaps, J. D., Forer, A. & Spurck, T. Traction fibre: toward a “tensegral” model of the spindle. Cell. Motil. Cytoskel. 37, 1–6 (1997).
Mitchison, T. J. et al. Roles of polymerization dynamics, opposed motors, and a tensile element in governing the length of Xenopus extract meiotic spindles. Mol. Biol. Cell 16, 3064–3076 (2005).
Qi, H. et al. East interacts with Megator and localizes to the putative spindle matrix during mitosis in Drosophila. J. Cell Biochem. 95, 1284–1291 (2005).
Qi, H. et al. Megator, an essential coiled-coil protein that localizes to the putative spindle matrix during mitosis in Drosophila. Mol. Biol. Cell 15, 4854–4865 (2004).
Rath, U. et al. Chromator, a novel and essential chromodomain protein interacts directly with the putative spindle matrix protein Skeletor. J. Cell Biochem. 93, 1033–1047 (2004).
Walker, D. L. et al. Skeletor, a novel chromosomal protein that redistributes during mitosis provides evidence for the formation of a spindle matrix. J. Cell Biol. 151, 1401–1411 (2000).
Lince-Faria, M. et al. Spatialtemporal control of mitosis by the conserved spindle matrix protein Megator. J. Cell Biol. 184, 647–657 (2009).
Ding, Y. et al. Chromator is required for proper microtubule spindle formation and mitosis in Drosophila. Dev. Biol. 334, 253–263 (2009).
Lee, S. H., Sterling, H., Burlingame, A. & McCormick, F. Tpr directly binds to Mad1 and Mad2 and is important for the Mad1-Mad2-mediated mitotic spindle checkpoint. Genes Dev. 22, 2926–2931 (2008).
Abad, P. C. et al. NuMA influences higher order chromatin organization in human mammary epithelium. Mol. Biol. Cell 18, 348–361 (2007).
Compton, D. & Cleveland, D. NuMA is required for the proper completion of mitosis. J. Cell Biol. 120, 947–957 (1993).
Compton, D. & Cleveland, D. NuMA, a nuclear protein involved in mitosis and nuclear reformation. Curr. Opin. Cell Biol. 6, 343–346 (1994).
Merdes, A., Ramyar, K., Vechio, J. & Cleveland, D. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell 87, 447–458 (1996).
Fant, X., Merdes, A. & Haren, L. Cell and molecular biology of spindle poles and NuMA. Int. Rev. Cytol. 238, 1–57 (2004).
Chang, P., Coughlin, M. & Mitchison, T. J. Interaction between Poly(ADP-ribose) and NuMA contributes to mitotic spindle pole assembly. Mol. Biol. Cell 20, 4575–4585 (2009).
Chang, P., Jacobson, M. K. & Mitchison, T. J. Poly(ADP-ribose) is required for spindle assembly and structure. Nature 432, 645–649 (2004).
Chang, P., Coughlin, M. & Mitchison, T. J. Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nature Cell Biol. 7, 1133–1139 (2005).
Dechat, T. et al. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 22, 832–853 (2008).
Beaudouin, J., Gerlich, D., Daigle, N., Eils, R. & Ellenberg, J. Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell 108, 83–96 (2002).
Salina, D. et al. Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell 108, 97–107 (2002).
Ma, L. et al. Requirement for Nudel and dynein for assembly of the lamin B spindle matrix. Nature Cell Biol. 11, 247–256 (2009).
Tsai, M.-Y. et al. A mitotic lamin B matrix induced by RanGTP required for spindle assembly. Science 311, 1887–1893 (2006).
Zheng, Y. & Tsai, M.-Y. The mitotic spindle matrix: a fibro-membranous lamin connection. Cell Cycle 5, 2345–2347 (2006).
Tsai, M.-Y. et al. A Ran signalling pathway mediated by the mitotic kinase Aurora A in spindle assembly. Nature Cell Biol. 5, 242–248 (2003).
Tsai, M.-Y. & Zheng, Y. Aurora A kinase-coated beads function as microtubule-organizing centers and enhance RanGTP-induced spindle assembly. Curr. Biol. 15, 2156–2163 (2005).
Orjalo, A. V. et al. The Nup107–160 nucleoporin complex is required for correct bipolar spindle assembly. Mol. Biol. Cell 17, 3806–3816 (2006).
Civelekoglu-Scholey, G., Tao, L., Brust-Mascher, I., Wollman, R. & Scholey, J. M. Prometaphase spindle maintenance by an antagonistic motor-dependent force balance made robust by a disassembling lamin-B envelope. J. Cell Biol. 188, 49–68 (2010).
Cao, K., Nakajima, R., Meyer, H. H. & Zheng, Y. The AAA-ATPase Cdc48/p97 regulates spindle disassembly at the end of mitosis. Cell 115, 355–367 (2003).
Cao, K. & Zheng, Y. The Cdc48/p97-Ufd1-Npl4 complex: its potential role in coordinating cellular morphogenesis during M-G1 transition. Cell Cycle 3, 422–424 (2004).
Royle, S. J., Bright, N. A. & Lagnado, L. Clathrin is required for the function of the mitotic spindle. Nature 434, 1152–1157 (2005).
Sutterlin, C., Polishchuk, R., Pecot, M. & Malhotra, V. The Golgi-associated protein GRASP65 regulates spindle dynamics and is essential for cell division. Mol. Biol. Cell 16, 3211–3222 (2005).
Vong, Q. P., Cao, K., li, H. Y., Iglesias, P. A. & Zheng, Y. Chromosome alignment and segregation regulated by ubiquitination of Surivin. Science 310, 1499–1504 (2005).
Lehtonen, S. et al. The endocytic adaptor protein ARH associates with motor and centrosomal proteins and is involved in centrosome assembly and cytokinesis. Mol. Biol. Cell 19, 2949–2961 (2008).
Boucrot, E. & Kirchhausen, T. Endosomal recycling controls plasma membrane area during mitosis. Proc. Natl Acad. Sci. USA 104, 7939–7944 (2007).
Liu, Y. et al. The Sac1 phosphoinositide phosphatase regulates golgi membrane morphology and mitotic spindle organization in mammals. Mol. Biol. Cell 19, 3080–3096 (2008).
Liu, Z. & Zheng, Y. A requirement for epsin in mitotic membrane and spindle organization. J. Cell Biol. 186, 473–480 (2009).
Ducat, D. C., Kawaguchi, S., Liu, H., Yates, J., 3rd & Zheng, Y. Regulation of microtubule assembly and organization in mitosis by the AAA+ ATPase Pontin. Mol. Biol. Cell 19, 3097–3110 (2008).
Yokoyama, H., Rybina, S., Santarella-Mellwig, R., Mattaj, I. W. & Karsenti, E. ISWI is a RanGTP-dependent MAP required for chromosome segregation. J. Cell Biol. 187, 813–829 (2009).
Zhang, C. & Clarke, P. R. Chromatin-independent nuclear envelope assembly induced by Ran GTPase in Xenopus egg extracts. Science 288, 1429–1432 (2000).
Hetzer, M., Bilbao-Cortes, D., Walther, T. C., Gruss, O. J. & Mattaj, I. W. GTP hydrolysis by Ran is required for nuclear envelope assembly. Mol. Cell 5, 1013–1024 (2000).
Moore, J. K., Stuchell-Brereton, M. D. & Cooper, J. A. Function of dynein in budding yeast: mitotic spindle position in a polarized cell. Cell. Motil. Cytoskel. 66, 546–555 (2009).
Gonzalez, Y. et al. Nuclear shape, growth and integrity in the closed mitosis of fission yeast depends on the Ran-GTPase system, the spindle pole body and the endoplasmic reticulum. J. Cell Sci. 122, 2464–2472 (2009).
De Souza, C. P., Hashmi, S. B., Nayak, T., Oakley, B. & Osmani, S. A. Mlp1 acts as a mitotic scaffold to spatially regulate spindle assembly checkpoint proteins in Aspergillus nidulans. Mol. Biol. Cell 20, 2146–2159 (2009).
Shcheprova, Z., Baldi, S., Frei, S. B., Gonnet, G. & Barrel, Y. A mechanism for asymmetric segregation of age during yeast budding. Nature 454, 728–734 (2008).
Siller, K. H. & Doe, C. O. Spindle orientation during assymetric cell division. Nature Cell Biol. 11, 365–374 (2009).
Wilde, A. & Zheng, Y. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science 284, 1359–1362 (1999).
Kalab, P., Pu, R. & Dasso, M. The Ran GTPase regulates mitotic spindle assembly. Curr. Biol. 9, 481–484 (1999).
Ohba, T., Nakamura, M., Nishitani, H. & Nishimoto, T. Self-organization of microtubule asters induced in Xenopus egg extracts by GTP-bound Ran. Science 284, 1356–1358 (1999).
Carazo-Salas, R. E. et al. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature 400, 178–181 (1999).
Li, H., Wirtz, D. & Zheng, Y. A mechanism of coupling RCC1 mobility to RanGTP production on the chromatin in vivo. J. Cell Biol. 160, 635–644 (2003).
Li, H. Y. & Zheng, Y. Phosphorylation of RCC1 in mitosis is essential for producing a high RanGTP concentration on chromosomes and for spindle assembly in mammalian cells. Genes Dev. 18, 512–527 (2004).
Kalab, P., Pralle, A., Isacoff, E. Y., Heald, R. & Weis, K. Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature 440, 697–701 (2006).
Walczak, C. E. & Heald, R. Mechanism of mitotic spindle assembly and function. Int. Rev. Cytol. 265, 111–158 (2008).
Acknowledgements
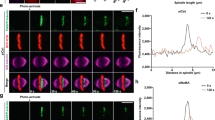
I thank B. Goodman for spindle drawings and B. Guo, K. Jung and members of the Zheng laboratory for helpful comments. I apologize to colleagues whose work could not be cited owing to space limitations. The work on mitosis in the Y.Z. laboratory is supported by the National Institute of General Medical Sciences (GM056312). Y.Z. is an investigator of the Howard Hughes Medical Institute. The image in the middle panel of figure 3 part c is courtesy of L. Ma.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The author declares no competing financial interests.
Related links
Rights and permissions
About this article
Cite this article
Zheng, Y. A membranous spindle matrix orchestrates cell division. Nat Rev Mol Cell Biol 11, 529–535 (2010). https://doi.org/10.1038/nrm2919
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm2919
This article is cited by
-
Liquid-liquid phase separation in biology: mechanisms, physiological functions and human diseases
Science China Life Sciences (2020)
-
Bidirectional motility of kinesin-5 motor proteins: structural determinants, cumulative functions and physiological roles
Cellular and Molecular Life Sciences (2018)
-
Mitotic spindle assembly in animal cells: a fine balancing act
Nature Reviews Molecular Cell Biology (2017)
-
An intrinsic mechanism controls reactivation of neural stem cells by spindle matrix proteins
Nature Communications (2017)
-
Interpolar microtubules are dispensable in fission yeast meiosis II
Nature Communications (2012)