Key Points
-
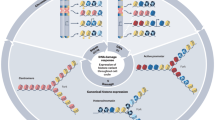
Chromatin is organized into specialized domains, some of which contain specialized histones called variant histones. Variant histones have evolved to carry out functions that are distinct from those of the major core histones.
-
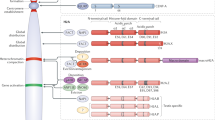
Histones are deposited onto DNA by chaperones. Recent studies have shown that some chaperones recruit specific histone variants for deposition. The chaperones HIRA and SWR1 specifically recruit and exchange the histone variants H3.3 and H2AZ, respectively, whereas the chromatin assembly factor-1 (CAF1) mediates the deposition of the H3.1 major core histone in a process that is coupled to DNA replication.
-
The implications of two modes of deposition of the histones H3 and H4 (that is, tetramers versus dimers) are discussed, as they are important for the transmission of epigenetic information from the mother to the daughter cells.
-
The functions and possible modes of deposition of other variant histones such as CENPA, H2AX and others are also discussed.
-
Both H3.3 and H2AZ have been suggested to function in activated transcription on the basis of their localization to euchromatic loci. Models are proposed whereby histone exchangers coordinate with FACT — a chaperone that has a crucial role in facilitating transcription elongation on chromatin — to allow the incorporation of histone variants during the process of transcription.
-
A model is also presented that addresses CAF1-mediated deposition of histone H3 that is methylated at Lys9, through CAF1 interaction with the histone methyltransferase SUV39H1, which results in the formation of repressive chromatin.
Abstract
A fascinating aspect of how chromatin structure impacts on gene expression and cellular identity is the transmission of information from mother to daughter cells, independently of the primary DNA sequence. This epigenetic information seems to be contained within the covalent modifications of histone polypeptides and the distinctive characteristics of variant histone subspecies. There are specific deposition pathways for some histone variants, which provide invaluable mechanistic insights into processes whereby the major histones are exchanged for their more specialized counterparts.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Malik, H. S. & Henikoff, S. Phylogenomics of the nucleosome. Nature Struct. Biol. 10, 882–891 (2003). An outstanding, comprehensive review on the evolution and functions of variant histones.
Kimmins, S. S. & Sassone-Corsi, P. Chromatin remodelling and epigenetic features of germ cells. Nature (in the press).
Jackson, V. In vivo studies on the dynamics of histone–DNA interaction: evidence for nucleosome dissolution during replication and transcription and a low level of dissolution independent of both. Biochemistry 29, 719–731 (1990).
Kimura, H. & Cook, P. R. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 153, 1341–1353 (2001).
Lever, M. A., Th'ng, J. P., Sun, X. & Hendzel, M. J. Rapid exchange of histone H1.1 on chromatin in living human cells. Nature 408, 873–876 (2000).
Wilhelm, F. X., Wilhelm, M. L., Erard, M. & Duane, M. P. Reconstitution of chromatin: assembly of the nucleosome. Nucleic Acids Res. 5, 505–521 (1978).
Smith, S. & Stillman, B. Stepwise assembly of chromatin during DNA replication in vitro. EMBO J. 10, 971–980 (1991).
Sobel, R. E., Cook, R. G., Perry, C. A., Annunziato, A. T. & Allis, C. D. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc. Natl Acad. Sci. USA 92, 1237–1241 (1995).
Mello, J. A. & Almouzni, G. The ins and outs of nucleosome assembly. Curr. Opin. Genet. Dev. 11, 136–141 (2001).
Ma, X. J., Wu, J., Altheim, B. A., Schultz, M. C. & Grunstein, M. Deposition-related sites K5/K12 in histone H4 are not required for nucleosome deposition in yeast. Proc. Natl Acad. Sci. USA 95, 6693–6698 (1998).
Shibahara, K., Verreault, A. & Stillman, B. The N-terminal domains of histones H3 and H4 are not necessary for chromatin assembly factor-1-mediated nucleosome assembly onto replicated DNA in vitro. Proc. Natl Acad. Sci. USA 97, 7766–7771 (2000).
Tyler, J. K. et al. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature 402, 555–560 (1999).
Nishioka, K. et al. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol. Cell 9, 1201–1213 (2002).
Rice, J. C. et al. Mitotic-specific methylation of histone H4 Lys 20 follows increased PR-Set7 expression and its localization to mitotic chromosomes. Genes Dev. 16, 2225–2230 (2002).
Bracken, A. P. et al. EZH2 is downstream of the pRB–E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 22, 5323–5335 (2003).
Loyola, A. & Almouzni, G. Histone chaperones, a supporting role in the limelight. Biochim. Biophys. Acta 1677, 3–11 (2004).
Zalensky, A. O. et al. Human testis/sperm-specific histone H2B (hTSH2B). Molecular cloning and characterization. J. Biol. Chem. 277, 43474–43480 (2002).
Churikov, D. et al. Novel human testis-specific histone H2B encoded by the interrupted gene on the X chromosome. Genomics 84, 745–756 (2004).
Ahmad, K. & Henikoff, S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9, 1191–1200 (2002). Shows the direct visualization of RI incorporation of H3.3 into chromatin.
McKittrick, E., Gafken, P. R., Ahmad, K. & Henikoff, S. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc. Natl Acad. Sci. USA 101, 1525–1530 (2004).
Tagami, H., Ray-Gallet, D., Almouzni, G. & Nakatani, Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116, 51–61 (2004). Describes the existence of unique pre-deposition complexes that mediate the incorporation of histones H3.1 and H3.3.
Adkins, M. W. & Tyler, J. K. The histone chaperone Asf1p mediates global chromatin disassembly in vivo. J. Biol. Chem. 279, 52069–52074 (2004).
Maga, G. & Hubscher, U. Proliferating cell nuclear antigen (PCNA): a dancer with many partners. J. Cell Sci. 116, 3051–3060 (2003).
Moggs, J. G. et al. A CAF-1–PCNA-mediated chromatin assembly pathway triggered by sensing DNA damage. Mol. Cell. Biol. 20, 1206–1218 (2000).
Shibahara, K. & Stillman, B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell 96, 575–585 (1999).
Ray-Gallet, D. et al. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol. Cell 9, 1091–1100 (2002).
Mello, J. A. et al. Human Asf1 and CAF-1 interact and synergize in a repair-coupled nucleosome assembly pathway. EMBO Rep. 3, 329–334 (2002).
Palmer, D. K., O'Day, K., Wener, M. H., Andrews, B. S. & Margolis, R. L. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J. Cell Biol. 104, 805–815 (1987).
Sullivan, K. F., Hechenberger, M. & Masri, K. Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J. Cell Biol. 127, 581–592 (1994).
Howman, E. V. et al. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc. Natl Acad. Sci. USA 97, 1148–1153 (2000).
Sharp, J. A., Franco, A. A., Osley, M. A. & Kaufman, P. D. Chromatin assembly factor I and Hir proteins contribute to building functional kinetochores in S. cerevisiae. Genes Dev. 16, 85–100 (2002).
Wieland, G., Orthaus, S., Ohndorf, S., Diekmann, S. & Hemmerich, P. Functional complementation of human centromere protein A (CENP-A) by Cse4p from Saccharomyces cerevisiae. Mol. Cell. Biol. 24, 6620–6630 (2004).
Black, B. E. et al. Structural determinants for generating centromeric chromatin. Nature 430, 578–582 (2004).
Faast, R. et al. Histone variant H2A.Z is required for early mammalian development. Curr. Biol. 11, 1183–1187 (2001).
Clarkson, M. J., Wells, J. R., Gibson, F., Saint, R. & Tremethick, D. J. Regions of variant histone His2AvD required for Drosophila development. Nature 399, 694–697 (1999).
Allis, C. D. et al. hv1 is an evolutionarily conserved H2A variant that is preferentially associated with active genes. J. Biol. Chem. 261, 1941–1948 (1986).
Meneghini, M. D., Wu, M. & Madhani, H. D. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112, 725–736 (2003).
Rusche, L. N., Kirchmaier, A. L. & Rine, J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72, 481–516 (2003).
Suto, R. K., Clarkson, M. J., Tremethick, D. J. & Luger, K. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nature Struct. Biol. 7, 1121–1124 (2000).
Rangasamy, D., Berven, L., Ridgway, P. & Tremethick, D. J. Pericentric heterochromatin becomes enriched with H2A.Z during early mammalian development. EMBO J. 22, 1599–1607 (2003).
Rangasamy, D., Greaves, I. & Tremethick, D. J. RNA interference demonstrates a novel role for H2A.Z in chromosome segregation. Nature Struct. Mol. Biol. 11, 650–655 (2004).
Mizuguchi, G. et al. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303, 343–348 (2004). Describes the purification of the Swr1 chromatin-remodelling complex that resulted in the recognition of its role in the exchange of H2AZ into chromatin.
Krogan, N. J. et al. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell 12, 1565–1576 (2003). Reports the identification of the H2AZ exchange complex Swr1 by a genetic screen that was designed to identify novel genes involved in transcription elongation.
Kobor, M. S. et al. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2, E131 (2004). Describes the identification of the Swr1 histone-exchange complex by the biochemical analysis of the interaction partners of H2AZ that facilitate its incorporation into chromatin.
Matangkasombut, O., Buratowski, R. M., Swilling, N. W. & Buratowski, S. Bromodomain factor 1 corresponds to a missing piece of yeast TFIID. Genes Dev. 14, 951–962 (2000).
Ebbert, R., Birkmann, A. & Schuller, H. J. The product of the SNF2/SWI2 paralogue INO80 of Saccharomyces cerevisiae required for efficient expression of various yeast structural genes is part of a high-molecular-weight protein complex. Mol. Microbiol. 32, 741–751 (1999).
Park, Y. J., Dyer, P. N., Tremethick, D. J. & Luger, K. A new fluorescence resonance energy transfer approach demonstrates that the histone variant H2AZ stabilizes the histone octamer within the nucleosome. J. Biol. Chem. 279, 24274–24282 (2004).
Fan, J. Y., Gordon, F., Luger, K., Hansen, J. C. & Tremethick, D. J. The essential histone variant H2A.Z regulates the equilibrium between different chromatin conformational states. Nature Struct. Biol. 9, 172–176 (2002).
Belotserkovskaya, R. et al. FACT facilitates transcription-dependent nucleosome alteration. Science 301, 1090–1093 (2003).
Orphanides, G., LeRoy, G., Chang, C. H., Luse, D. S. & Reinberg, D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell 92, 105–116 (1998).
Downs, J. A., Lowndes, N. F. & Jackson, S. P. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature 408, 1001–1004 (2000).
Rogakou, E. P., Pilch, D. R., Orr, A. H., Ivanova, V. S. & Bonner, W. M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273, 5858–5868 (1998).
Stiff, T. et al. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 64, 2390–2396 (2004).
Madigan, J. P., Chotkowski, H. L. & Glaser, R. L. DNA double-strand break-induced phosphorylation of Drosophila histone variant H2Av helps prevent radiation-induced apoptosis. Nucleic Acids Res. 30, 3698–3705 (2002).
Celeste, A. et al. Genomic instability in mice lacking histone H2AX. Science 296, 922–927 (2002).
Celeste, A. et al. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nature Cell Biol. 5, 675–679 (2003).
Fernandez-Capetillo, O. et al. H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Dev. Cell 4, 497–508 (2003).
Cheung, W. L. et al. Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase. Cell 113, 507–517 (2003).
Fernandez-Capetillo, O., Allis, C. D. & Nussenzweig, A. Phosphorylation of histone H2B at DNA double-strand breaks. J. Exp. Med. 199, 1671–1677 (2004).
de la Barre, A. E., Angelov, D., Molla, A. & Dimitrov, S. The N-terminus of histone H2B, but not that of histone H3 or its phosphorylation, is essential for chromosome condensation. EMBO J. 20, 6383–6393 (2001).
Fernandez-Capetillo, O., Lee, A., Nussenzweig, M. & Nussenzweig, A. H2AX: the histone guardian of the genome. DNA Repair (Amst.) 3, 959–967 (2004).
Pehrson, J. R. & Fried, V. A. MacroH2A, a core histone containing a large nonhistone region. Science 257, 1398–1400 (1992).
Costanzi, C. & Pehrson, J. R. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature 393, 599–601 (1998).
Mermoud, J. E., Costanzi, C., Pehrson, J. R. & Brockdorff, N. Histone macroH2A1.2 relocates to the inactive X chromosome after initiation and propagation of X-inactivation. J. Cell Biol. 147, 1399–1408 (1999).
Rasmussen, T. P., Mastrangelo, M. A., Eden, A., Pehrson, J. R. & Jaenisch, R. Dynamic relocalization of histone MacroH2A1 from centrosomes to inactive X chromosomes during X inactivation. J. Cell Biol. 150, 1189–1198 (2000).
Angelov, D. et al. The histone variant macroH2A interferes with transcription factor binding and SWI/SNF nucleosome remodeling. Mol. Cell 11, 1033–1041 (2003).
Chadwick, B. P., Valley, C. M. & Willard, H. F. Histone variant macroH2A contains two distinct macrochromatin domains capable of directing macroH2A to the inactive X chromosome. Nucleic Acids Res. 29, 2699–2705 (2001).
Gautier, T. et al. Histone variant H2ABbd confers lower stability to the nucleosome. EMBO Rep. 5, 715–720 (2004).
Bao, Y. et al. Nucleosomes containing the histone variant H2A.Bbd organize only 118 base pairs of DNA. EMBO J. 23, 3314–3324 (2004).
Bruno, M. et al. Histone H2A/H2B dimer exchange by ATP-dependent chromatin remodeling activities. Mol. Cell 12, 1599–1606 (2003).
Hirschhorn, J. N., Brown, S. A., Clark, C. D. & Winston, F. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 6, 2288–2298 (1992).
Mason, P. B. & Struhl, K. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol. Cell. Biol. 23, 8323–8333 (2003).
Saunders, A. et al. Tracking FACT and the RNA polymerase II elongation complex through chromatin in vivo. Science 301, 1094–1096 (2003).
Endoh, M. et al. Human Spt6 stimulates transcription elongation by RNA polymerase II in vitro. Mol. Cell. Biol. 24, 3324–3336 (2004).
Wada, T. et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 12, 343–356 (1998).
Hartzog, G. A., Wada, T., Handa, H. & Winston, F. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 12, 357–369 (1998).
Kaplan, C. D., Morris, J. R., Wu, C. & Winston, F. Spt5 and spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster. Genes Dev. 14, 2623–2634 (2000).
Andrulis, E. D., Guzman, E., Doring, P., Werner, J. & Lis, J. T. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev. 14, 2635–2649 (2000).
Bortvin, A. & Winston, F. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science 272, 1473–1476 (1996).
Kaplan, C. D., Laprade, L. & Winston, F. Transcription elongation factors repress transcription initiation from cryptic sites. Science 301, 1096–1099 (2003).
Murzina, N., Verreault, A., Laue, E. & Stillman, B. Heterochromatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins. Mol. Cell 4, 529–540 (1999).
Yamamoto, K. & Sonoda, M. Self-interaction of heterochromatin protein 1 is required for direct binding to histone methyltransferase, SUV39H1. Biochem. Biophys. Res. Commun. 301, 287–292 (2003).
Quivy, J. P. et al. A CAF-1 dependent pool of HP1 during heterochromatin duplication. EMBO J. 23, 3516–3526 (2004).
Rice, J. C. et al. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol. Cell 12, 1591–1598 (2003).
Peters, A. H. et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell 12, 1577–1589 (2003).
Schotta, G. et al. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 18, 1251–1262 (2004).
Heard, E. et al. Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell 107, 727–738 (2001).
Silva, J. et al. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed–Enx1 polycomb group complexes. Dev. Cell 4, 481–495 (2003).
Plath, K. et al. Role of histone H3 lysine 27 methylation in X inactivation. Science 300, 131–135 (2003).
Kohlmaier, A. et al. A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol. 2, E171 (2004).
Vaquero, A., Loyola, A. & Reinberg, D. The constantly changing face of chromatin. Sci. Aging Knowledge Environ. 2003, RE4 (2003).
Zhang, Y. & Reinberg, D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 15, 2343–2360 (2001).
Turner, B. M. Cellular memory and the histone code. Cell 111, 285–291 (2002).
Jenuwein, T. & Allis, C. D. Translating the histone code. Science 293, 1074–1080 (2001).
Cuthbert, G. L. et al. Histone deimination antagonizes arginine methylation. Cell 118, 545–553 (2004).
Wang, H. et al. mAM facilitates conversion by ESET of dimethyl to trimethyl lysine 9 of histone H3 to cause transcriptional repression. Mol. Cell 12, 475–487 (2003).
Shi, Y. et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119, 941–953 (2004).
Acknowledgements
We thank Ken Marians, Jerry Hurwitz, Steven Henikoff, Yoshihiro Nakatani and Geneviève Almouzni for helpful discussions. We would also like to thank Lynne Vales for critical reading of the manuscript and members of the Reinberg laboratory for discussions. We apologize to colleagues whose work we have not cited owing to space limitations. This work was supported by grants from the National Institutes of Health and the Howard Hughes Medical Institute to D.R.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- ORPHAN GENE
-
A protein-coding region that bears little or no homology to genes in distant species.
- HISTONE LYSINE METHYLTRANSFERASE
-
An enzyme that catalyses the transfer of methyl groups onto the ε-amino residue of lysines in histones.
- ALPHA SATELLITE REPEAT
-
Large highly repetitive stretches of (A+T)-rich DNA sequences in the human genome that are usually untranscribed.
- HISTONE CHAPERONE
-
A protein that escorts histones to DNA for deposition.
- CHROMATIN-REMODELLING FACTOR
-
A protein that alters the dynamic organization of nucleosomes to help in the activation or repression of gene expression.
- HISTONE ACETYLTRANSFERASE
-
An enzyme that catalyses the addition of an acetyl group to specific lysine residues in histones.
- MACRONUCLEUS
-
The larger of the two nuclei in the unicellular ciliate Tetrahymena thermophila. This is the somatic nucleus and is transcriptionally active.
- MICRONUCLEUS
-
The smaller 'germline' nucleus in Tetrahymena thermophila, which is transcriptionally silent.
- CONJUGATION
-
A process of sexual reproduction that occurs in some unicellular organisms and that involves the exchange of genetic material between two cells through a so-called sex pilus.
- MATING-TYPE LOCUS
-
The genomic region in yeast that determines the mating type or 'sex' of the haploid yeast cell.
- BROMODOMAIN
-
An evolutionarily conserved domain that has been shown to bind to acetylated residues.
- CHROMATIN IMMUNOPRECIPITATION
-
(ChIP). A technique by which direct or indirect protein–DNA interactions in chromatin can be studied using antibodies against specific chromosomal proteins.
- X INACTIVATION
-
The process whereby one of the two copies of the X chromosome in female mammals is silenced to compensate for the presence of a single copy in males.
- LEUCINE-ZIPPER MOTIF
-
A leucine-rich protein domain that mediates interactions with other proteins with a similar domain.
Rights and permissions
About this article
Cite this article
Sarma, K., Reinberg, D. Histone variants meet their match. Nat Rev Mol Cell Biol 6, 139–149 (2005). https://doi.org/10.1038/nrm1567
Issue Date:
DOI: https://doi.org/10.1038/nrm1567
This article is cited by
-
The patterns and participants of parental histone recycling during DNA replication in Saccharomyces cerevisiae
Science China Life Sciences (2023)
-
Nucleosomes positioning around transcriptional start site of tumor suppressor (Rbl2/p130) gene in breast cancer
Molecular Biology Reports (2018)
-
The histone variant H2A.X is a regulator of the epithelial–mesenchymal transition
Nature Communications (2016)
-
Prospects for the development of epigenetic drugs for CNS conditions
Nature Reviews Drug Discovery (2015)