Key Points
-
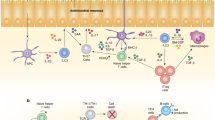
T-bet is expressed in many different cell types of the innate and adaptive immune system in both myeloid and lymphoid lineages.
-
T-bet expression arose early in evolution, before the appearance of the adaptive immune system, which suggests that its function in B cells and T cells may partly reflect coopted transcriptional pathways.
-
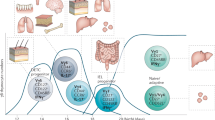
T-bet has a crucial role in regulating mucosal homeostasis, mainly via its function in dendritic cells and innate lymphoid cells.
-
T-bet regulates the T helper 1 (TH1) cell differentiation programme by recruiting chromatin-modifying enzymes, which promote permissive chromatin marks at TH1 cell-specific loci by directly regulating the expression of interferon-γ (Ifng) and approximately 27 TH1 cell-specific genes, and by organizing the three-dimensional architecture of the Ifng locus.
-
T-bet blocks the differentiation of other CD4+ TH cell subsets either by inhibiting the expression of TH cell lineage-specifying transcription factors in TH precursor cells or by interfering with their transcriptional activity.
-
T-bet expression in other fully differentiated TH cell subsets results in the acquisition of the TH1 cell phenotype, which may or may not be accompanied by the repression of the existing gene expression profile.
-
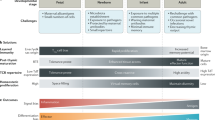
During acute infections, T-bet balances terminal differentiation and memory cell potential in both CD4+ and CD8+ T cells. Its expression correlates with the terminal differentiation of CD4+ and CD8+ T effector cells, and its absence correlates with higher memory cell potential.
-
During chronic infections,T-bet expression in CD8+ T cells prevents cell exhaustion.
Abstract
Originally described over a decade ago as a T cell transcription factor regulating T helper 1 cell lineage commitment, T-bet is now recognized as having an important role in many cells of the adaptive and innate immune system. T-bet has a fundamental role in coordinating type 1 immune responses by controlling a network of genetic programmes that regulate the development of certain immune cells and the effector functions of others. Many of these transcriptional networks are conserved across innate and adaptive immune cells and these shared mechanisms highlight the biological functions that are regulated by T-bet.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Flajnik, M. F. & Kasahara, M. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nature Rev. Genet. 11, 47–59 (2010).
Guo, P. et al. Dual nature of the adaptive immune system in lampreys. Nature 459, 796–801 (2009).
Horton, A. C. & Gibson-Brown, J. J. Evolution of developmental functions by the Eomesodermin, T-brain-1, Tbx21 subfamily of T-box genes: insights from amphioxus. J. Exp. Zool. 294, 112–121 (2002).
Kanhere, A. et al. T-bet and GATA3 orchestrate Th1 and Th2 differentiation through lineage-specific targeting of distal regulatory elements. Nature Commun. 3, 1268 (2012).
Vahedi, G. et al. STATs shape the active enhancer landscape of T cell populations. Cell 151, 981–993 (2012).
Ciofani, M. et al. A validated regulatory network for Th17 cell specification. Cell 151, 289–303 (2012).
Yagi, R. et al. The transcription factor GATA3 actively represses RUNX3 protein-regulated production of interferon-γ. Immunity 32, 507–517 (2010).
Zhu, J. et al. The transcription factor T-bet is induced by multiple pathways and prevents an endogenous Th2 cell program during Th1 cell responses. Immunity 37, 660–673 (2012). This is the first description of T-bet-specific reporter mice that can be used to address important questions about the regulation of T-bet expression during T H 1 cell differentiation.
Wei, G. et al. Genome-wide analyses of transcription factor GATA3-mediated gene regulation in distinct T cell types. Immunity 35, 299–311 (2011).
Whyte, W. A. et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013).
Lighvani, A. A. et al. T-bet is rapidly induced by interferon-γ in lymphoid and myeloid cells. Proc. Natl Acad. Sci. USA 98, 15137–15142 (2001).
Lugo-Villarino, G., Maldonado-Lopez, R., Possemato, R., Penaranda, C. & Glimcher, L. H. T-bet is required for optimal production of IFN-γ and antigen-specific T cell activation by dendritic cells. Proc. Natl Acad. Sci. USA 100, 7749–7754 (2003).
Lugo-Villarino, G., Ito, S., Klinman, D. M. & Glimcher, L. H. The adjuvant activity of CpG DNA requires T-bet expression in dendritic cells. Proc. Natl Acad. Sci. USA 102, 13248–13253 (2005).
Wang, J. et al. Transcription factor T-bet regulates inflammatory arthritis through its function in dendritic cells. J. Clin. Invest. 116, 414–421 (2006).
Powell, N., Canavan, J. B., MacDonald, T. T. & Lord, G. M. Transcriptional regulation of the mucosal immune system mediated by T-bet. Mucosal Immunol. 3, 567–577 (2010).
Alcaide, P. et al. Dendritic cell expression of the transcription factor T-bet regulates mast cell progenitor homing to mucosal tissue. J. Exp. Med. 204, 431–439 (2007).
Garrett, W. S. et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell 131, 33–45 (2007). This paper describes the microbiome-dependent spontaneous colitis that occurs in the absence of T-bet and RAG2 as a result of the derepression of TNF in mucosal DCs.
Garrett, W. S. et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe 8, 292–300 (2010).
Garrett, W. S. et al. Colitis-associated colorectal cancer driven by T-bet deficiency in dendritic cells. Cancer Cell 16, 208–219 (2009).
Hecht, G. A. Inflammatory bowel disease — live transmission. N. Engl. J. Med. 358, 528–530 (2008).
Jenner, R. G. et al. The transcription factors T-bet and GATA-3 control alternative pathways of T-cell differentiation through a shared set of target genes. Proc. Natl Acad. Sci. USA 106, 17876–17881 (2009).
Powell, N. et al. The transcription factor T-bet regulates intestinal inflammation mediated by interleukin-7 receptor+ innate lymphoid cells. Immunity 37, 674–684 (2012). This is the first description of a functional role of T-bet that is expressed in ILCs in the regulation of mucosal immunity.
Spits, H. & Cupedo, T. Innate lymphoid cells: emerging insights in development, lineage relationships, and function. Annu. Rev. Immunol. 30, 647–675 (2012).
Bernink, J. H. et al. Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nature Immunol. 14, 221–229 (2013). This is the first description of human ILCs expressing T-bet that correlate with mucosal inflammation in Crohn's disease.
Fuchs, A. et al. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-γ-producing cells. Immunity 38, 769–781 (2013).
Spits, H. et al. Innate lymphoid cells — a proposal for uniform nomenclature. Nature Rev. Immunol. 13, 145–149 (2013).
Vonarbourg, C. et al. Regulated expression of nuclear receptor RORγt confers distinct functional fates to NK cell receptor-expressing RORγt+ innate lymphocytes. Immunity 33, 736–751 (2010).
Sciume, G. et al. Distinct requirements for T-bet in gut innate lymphoid cells. J. Exp. Med. 209, 2331–2338 (2012).
Klose, C. S. et al. A T-bet gradient controls the fate and function of CCR6-RORγt+ innate lymphoid cells. Nature 494, 261–265 (2013). This paper describes the mechanistic relationship of T-bet-expressing ILCs with RORγt and CCR6 expression and how this affects mucosal immunity.
Gordon, S. M. et al. The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity 36, 55–67 (2012).
Townsend, M. J. et al. T-bet regulates the terminal maturation and homeostasis of NK and Vα14i NKT cells. Immunity 20, 477–494 (2004). This paper is the first to describe the function of T-bet in NK and iNKT cells.
Werneck, M. B., Lugo-Villarino, G., Hwang, E. S., Cantor, H. & Glimcher, L. H. T-bet plays a key role in NK-mediated control of melanoma metastatic disease. J. Immunol. 180, 8004–8010 (2008).
Matsuda, J. L. et al. T-bet concomitantly controls migration, survival, and effector functions during the development of Vα14i NKT cells. Blood 107, 2797–2805 (2006).
Kim, H. Y. et al. The development of airway hyperreactivity in T-bet-deficient mice requires CD1d-restricted NKT cells. J. Immunol. 182, 3252–3261 (2009).
Yin, Z. et al. T-bet expression and failure of GATA-3 cross-regulation lead to default production of IFN-γ by γδ T cells. J. Immunol. 168, 1566–1571 (2002).
Chen, L. et al. Epigenetic and transcriptional programs lead to default IFN-γ production by γδ T cells. J. Immunol. 178, 2730–2736 (2007).
Szabo, S. J. et al. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell 100, 655–669 (2000). This is the first report to show the role of T-bet as the master regulator of the T H 1 cell differentiation programme.
Lazarevic, V. et al. T-bet represses TH17 differentiation by preventing Runx1-mediated activation of the gene encoding RORγt. Nature Immunol. 12, 96–104 (2011).
Mathur, A. N. et al. T-bet is a critical determinant in the instability of the IL-17-secreting T-helper phenotype. Blood 108, 1595–1601 (2006).
Villarino, A. V., Gallo, E. & Abbas, A. K. STAT1-activating cytokines limit Th17 responses through both T-bet-dependent and -independent mechanisms. J. Immunol. 185, 6461–6471 (2010).
Ravindran, R., Foley, J., Stoklasek, T., Glimcher, L. H. & McSorley, S. J. Expression of T-bet by CD4 T cells is essential for resistance to Salmonella infection. J. Immunol. 175, 4603–4610 (2005).
Sullivan, B. M. et al. Increased susceptibility of mice lacking T-bet to infection with Mycobacterium tuberculosis correlates with increased IL-10 and decreased IFN-γ production. J. Immunol. 175, 4593–4602 (2005).
Szabo, S. J. et al. Distinct effects of T-bet in TH1 lineage commitment and IFN-γ production in CD4 and CD8 T cells. Science 295, 338–342 (2002).
Afkarian, M. et al. T-bet is a STAT1-induced regulator of IL-12R expression in naive CD4+ T cells. Nature Immunol. 3, 549–557 (2002).
Mullen, A. C. et al. Role of T-bet in commitment of TH1 cells before IL-12-dependent selection. Science 292, 1907–1910 (2001).
Schulz, E. G., Mariani, L., Radbruch, A. & Hofer, T. Sequential polarization and imprinting of type 1 T helper lymphocytes by interferon-γ and interleukin-12. Immunity 30, 673–683 (2009).
Thieu, V. T. et al. Signal transducer and activator of transcription 4 is required for the transcription factor T-bet to promote T helper 1 cell-fate determination. Immunity 29, 679–690 (2008).
Liao, W., Lin, J. X., Wang, L., Li, P. & Leonard, W. J. Modulation of cytokine receptors by IL-2 broadly regulates differentiation into helper T cell lineages. Nature Immunol. 12, 551–559 (2011).
Smith, K. M. et al. miR-29ab1 deficiency identifies a negative feedback loop controlling Th1 bias that is dysregulated in multiple sclerosis. J. Immunol. 189, 1567–1576 (2012).
Steiner, D. F. et al. MicroRNA-29 regulates T-box transcription factors and interferon-γ production in helper T cells. Immunity 35, 169–181 (2011).
Jang, E. J., Park, H. R., Hong, J. H. & Hwang, E. S. Lysine 313 of T-box is crucial for modulation of protein stability, DNA binding, and threonine phosphorylation of T-bet. J. Immunol. 190, 5764–5770 (2013).
Miller, S. A. & Weinmann, A. S. Molecular mechanisms by which T-bet regulates T-helper cell commitment. Immunol. Rev. 238, 233–246 (2010).
Miller, S. A., Mohn, S. E. & Weinmann, A. S. Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol. Cell 40, 594–605 (2010).
Miller, S. A., Huang, A. C., Miazgowicz, M. M., Brassil, M. M. & Weinmann, A. S. Coordinated but physically separable interaction with H3K27-demethylase and H3K4-methyltransferase activities are required for T-box protein-mediated activation of developmental gene expression. Genes Dev. 22, 2980–2993 (2008).
Avni, O. et al. TH cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nature Immunol. 3, 643–651 (2002).
Balasubramani, A. et al. Modular utilization of distal cis-regulatory elements controls Ifng gene expression in T cells activated by distinct stimuli. Immunity 33, 35–47 (2010).
Shnyreva, M. et al. Evolutionarily conserved sequence elements that positively regulate IFN-γ expression in T cells. Proc. Natl Acad. Sci. USA 101, 12622–12627 (2004).
Hatton, R. D. et al. A distal conserved sequence element controls Ifng gene expression by T cells and NK cells. Immunity 25, 717–729 (2006). This study provides detailed analysis of the regulatory elements within the Ifng locus and maps T-bet-dependent enhancers.
Mullen, A. C. et al. Hlx is induced by and genetically interacts with T-bet to promote heritable TH1 gene induction. Nature Immunol. 3, 652–658 (2002).
Djuretic, I. M. et al. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nature Immunol. 8, 145–153 (2007).
Sekimata, M. et al. CCCTC-binding factor and the transcription factor T-bet orchestrate T helper 1 cell-specific structure and function at the interferon-γ locus. Immunity 31, 551–564 (2009).
Lord, G. M. et al. T-bet is required for optimal proinflammatory CD4+ T-cell trafficking. Blood 106, 3432–3439 (2005).
Zheng, W. & Flavell, R. A. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell 89, 587–596 (1997).
Hwang, E. S., Szabo, S. J., Schwartzberg, P. L. & Glimcher, L. H. T helper cell fate specified by kinase-mediated interaction of T-bet with GATA-3. Science 307, 430–433 (2005). This paper describes T-bet-mediated inhibition of GATA3 function during T H 2 cell differentiation.
Hwang, E. S., Hong, J. H. & Glimcher, L. H. IL-2 production in developing Th1 cells is regulated by heterodimerization of RelA and T-bet and requires T-bet serine residue 508. J. Exp. Med. 202, 1289–1300 (2005).
Hegazy, A. N. et al. Interferons direct Th2 cell reprogramming to generate a stable GATA-3+T-bet+ cell subset with combined Th2 and Th1 cell functions. Immunity 32, 116–128 (2010).
Hirota, K. et al. Fate mapping of IL-17-producing T cells in inflammatory responses. Nature Immunol. 12, 255–263 (2011).
Lee, Y. et al. Induction and molecular signature of pathogenic TH17 cells. Nature Immunol. 13, 991–999 (2012).
Ghoreschi, K. et al. Generation of pathogenic TH17 cells in the absence of TGF-β signalling. Nature 467, 967–971 (2010).
Yang, Y. et al. T-bet is essential for encephalitogenicity of both Th1 and Th17 cells. J. Exp. Med. 206, 1549–1564 (2009).
Lee, Y. K. et al. Late developmental plasticity in the T helper 17 lineage. Immunity 30, 92–107 (2009).
Mukasa, R. et al. Epigenetic instability of cytokine and transcription factor gene loci underlies plasticity of the T helper 17 cell lineage. Immunity 32, 616–627 (2010).
Wei, G. et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity 30, 155–167 (2009). This study shows that the maintenance of master transcription factor loci in a bivalent epigenetic state, which is characterized by the presence of repressive and permissive marks, is the underlying mechanism for the functional plasticity of T H cells.
Nakayamada, S. et al. Early Th1 cell differentiation is marked by a Tfh cell-like transition. Immunity 35, 919–931 (2011).
Oestreich, K. J., Huang, A. C. & Weinmann, A. S. The lineage-defining factors T-bet and Bcl-6 collaborate to regulate Th1 gene expression patterns. J. Exp. Med. 208, 1001–1013 (2011).
Oestreich, K. J., Mohn, S. E. & Weinmann, A. S. Molecular mechanisms that control the expression and activity of Bcl-6 in TH1 cells to regulate flexibility with a TFH-like gene profile. Nature Immunol. 13, 405–411 (2012).
Lu, K. T. et al. Functional and epigenetic studies reveal multistep differentiation and plasticity of in vitro-generated and in vivo-derived follicular T helper cells. Immunity 35, 622–632 (2011).
Hall, A. O. et al. The cytokines interleukin 27 and interferon-gamma promote distinct Treg cell populations required to limit infection-induced pathology. Immunity 37, 511–523 (2012).
Koch, M. A. et al. T-bet+ Treg cells undergo abortive Th1 cell differentiation due to impaired expression of IL-12 receptor β2. Immunity 37, 501–510 (2012).
Koch, M. A. et al. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nature Immunol. 10, 595–602 (2009).
Sullivan, B. M., Juedes, A., Szabo, S. J., von Herrath, M. & Glimcher, L. H. Antigen-driven effector CD8 T cell function regulated by T-bet. Proc. Natl Acad. Sci. USA 100, 15818–15823 (2003).
Pearce, E. L. et al. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science 302, 1041–1043 (2003).
Intlekofer, A. M. et al. Anomalous type 17 response to viral infection by CD8+ T cells lacking T-bet and eomesodermin. Science 321, 408–411 (2008).
Cruz-Guilloty, F. et al. Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J. Exp. Med. 206, 51–59 (2009).
Takemoto, N., Intlekofer, A. M., Northrup, J. T., Wherry, E. J. & Reiner, S. L. Cutting edge: IL-12 inversely regulates T-bet and eomesodermin expression during pathogen-induced CD8+ T cell differentiation. J. Immunol. 177, 7515–7519 (2006).
Pipkin, M. E. et al. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity 32, 79–90 (2010).
Joshi, N. S. et al. Inflammation directs memory precursor and short-lived effector CD8+ T cell fates via the graded expression of T-bet transcription factor. Immunity 27, 281–295 (2007). This paper shows the importance of the T-bet expression levels in determining the terminal differentiation of CD8+ effector cells or in determining CD8+ memory cell formation.
Rao, R. R., Li, Q., Gubbels Bupp, M. R. & Shrikant, P. A. Transcription factor Foxo1 represses T-bet-mediated effector functions and promotes memory CD8+ T cell differentiation. Immunity 36, 374–387 (2012).
Rao, R. R., Li, Q., Odunsi, K. & Shrikant, P. A. The mTOR kinase determines effector versus memory CD8+ T cell fate by regulating the expression of transcription factors T-bet and Eomesodermin. Immunity 32, 67–78 (2010). This paper describes the importance of mTOR kinase in determining effector versus memory CD8+ T cell fate through its effects on the regulation of T-bet expression.
Chang, J. T. et al. Asymmetric proteasome segregation as a mechanism for unequal partitioning of the transcription factor T-bet during T lymphocyte division. Immunity 34, 492–504 (2011).
Joshi, N. S. et al. Increased numbers of preexisting memory CD8 T cells and decreased T-bet expression can restrain terminal differentiation of secondary effector and memory CD8 T cells. J. Immunol. 187, 4068–4076 (2011).
Banerjee, A. et al. Cutting edge: The transcription factor eomesodermin enables CD8+ T cells to compete for the memory cell niche. J. Immunol. 185, 4988–4992 (2010).
Intlekofer, A. M. et al. Requirement for T-bet in the aberrant differentiation of unhelped memory CD8+ T cells. J. Exp. Med. 204, 2015–2021 (2007).
Intlekofer, A. M. et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nature Immunol. 6, 1236–1244 (2005).
Virgin, H. W., Wherry, E. J. & Ahmed, R. Redefining chronic viral infection. Cell 138, 30–50 (2009).
Blackburn, S. D. et al. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nature Immunol. 10, 29–37 (2009).
Kao, C. et al. Transcription factor T-bet represses expression of the inhibitory receptor PD-1 and sustains virus-specific CD8+ T cell responses during chronic infection. Nature Immunol. 12, 663–671 (2011).
Paley, M. A. et al. Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science 338, 1220–1225 (2012).
Marshall, H. D. et al. Differential expression of Ly6C and T-bet distinguish effector and memory Th1 CD4+ cell properties during viral infection. Immunity 35, 633–646 (2011).
Hale, J. S. et al. Distinct memory CD4+ T cells with commitment to T follicular helper- and T helper 1-cell lineages are generated after acute viral infection. Immunity 38, 805–817 (2013).
Pepper, M., Pagan, A. J., Igyarto, B. Z., Taylor, J. J. & Jenkins, M. K. Opposing signals from the Bcl6 transcription factor and the interleukin-2 receptor generate T helper 1 central and effector memory cells. Immunity 35, 583–595 (2011).
Nguyen, H. V. et al. The Ets-1 transcription factor is required for Stat1-mediated T-bet expression and IgG2a class switching in mouse B cells. Blood 119, 4174–4181 (2012).
Peng, S. L., Szabo, S. J. & Glimcher, L. H. T-bet regulates IgG class switching and pathogenic autoantibody production. Proc. Natl Acad. Sci. USA 99, 5545–5550 (2002).
Xu, W. & Zhang, J. J. Stat1-dependent synergistic activation of T-bet for IgG2a production during early stage of B cell activation. J. Immunol. 175, 7419–7424 (2005).
Yoshimoto, T. et al. Induction of IgG2a class switching in B cells by IL-27. J. Immunol. 173, 2479–2485 (2004).
Wang, N. S. et al. Divergent transcriptional programming of class-specific B cell memory by T-bet and RORα. Nature Immunol. 13, 604–611 (2012).
Serre, K. et al. CD8 T cells induce T-bet-dependent migration toward CXCR3 ligands by differentiated B cells produced during responses to alum-protein vaccines. Blood 120, 4552–4559 (2012).
Acknowledgements
We would like to acknowledge support from grants awarded by the US National Institutes of Health (grants CA112663 and PO1NSO38037) to L.H.G.; the Irvington Institute to V.L.; and to G.M.L. the Medical Research Council, UK (grant G0802068), the Wellcome Trust, UK, (grant WT088747MA) and the British Heart Foundation, UK, (grant PG/12/36/29444). G.M.L. is also supported by the National Institute for Health Research (NIHR) Biomedical Research Centre based at Guy's and St Thomas' National Health Service (NHS) Foundation Trust and King's College London, UK. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
L.H.G. is on the Board of Directors and holds equity in Bristol–Myers Squibb, USA.
Related links
Glossary
- Experimental autoimmune encephalomyelitis
-
(EAE). A mouse model of multiple sclerosis. It can be induced by the active immunization of mice with central nervous system (CNS)-derived antigens emulsified in complete Freund's adjuvant (in the case of active EAE) or by adoptive transfer of activated, T cell receptor (TCR)-transgenic CD4+ T cells, in which the TCRs recognize a CNS-derived antigen (in the case of passive EAE).
- Scurfy mice
-
Mice with a spontaneous mutation in forkhead box P3 (Foxp3), which leads to a rapidly fatal lymphoproliferative disease, causing death by ∼4 weeks of age.
- Mamammalian target of rapamcyin
-
(mTOR). A serine/threonine protein kinase that regulates cell metabolism, proliferation, survival, protein synthesis and transcription. It is activated by T cell receptor signalling and is sustained by inflammatory cytokines such as interleukin-12.
- AMP-activated protein kinase
-
(AMPK). A negative regulator of mammalian target of rapamycin (mTOR) kinase activity. It is activated in response to cellular stress and ATP deprivation. It can be also activated by the pharmacological agent metformin.
- Affinity maturation
-
The process by which B cells produce antibodies that have an increased affinity for antigens during an immune response.
- Class-switch recombination
-
(CSR). The process by which B cells produce antibodies of different isotypes without changing the antigen specificity of the variable region.
Rights and permissions
About this article
Cite this article
Lazarevic, V., Glimcher, L. & Lord, G. T-bet: a bridge between innate and adaptive immunity. Nat Rev Immunol 13, 777–789 (2013). https://doi.org/10.1038/nri3536
Published:
Issue Date:
DOI: https://doi.org/10.1038/nri3536