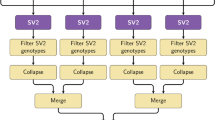
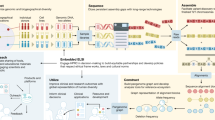
Key Points
-
The copy number variation (CNV) map of the human genome documents the extent and characteristics of CNV among healthy populations.
-
Depending on the level of stringency of the map, 4.8–9.7% of the human genome contributes to CNVs.
-
CNVs are distributed unevenly in the genome; the pericentromeric and subtelomeric regions of chromosomes show a particularly high rate of variation.
-
Various gene groups are affected differently by copy number variants. Genes that are associated with disease are the least affected by copy number variants, whereas paralogous genes have the most copy number variants.
-
More than 100 genes can be completely removed from the genome without producing apparent phenotypic consequences.
-
The CNV map will aid the interpretation of copy number variants of medical importance.
Abstract
A major contribution to the genome variability among individuals comes from deletions and duplications — collectively termed copy number variations (CNVs) — which alter the diploid status of DNA. These alterations may have no phenotypic effect, account for adaptive traits or can underlie disease. We have compiled published high-quality data on healthy individuals of various ethnicities to construct an updated CNV map of the human genome. Depending on the level of stringency of the map, we estimated that 4.8–9.5% of the genome contributes to CNV and found approximately 100 genes that can be completely deleted without producing apparent phenotypic consequences. This map will aid the interpretation of new CNV findings for both clinical and research applications.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Feuk, L., Carson, A. R. & Scherer, S. W. Structural variation in the human genome. Nature Rev. Genet. 7, 85–97 (2006). This is a comprehensive review of CNV and structural variation that suggests nomenclature for the newly emerging field.
Beckmann, J. S., Estivill, X. & Antonarakis, S. E. Copy number variants and genetic traits: closer to the resolution of phenotypic to genotypic variability. Nature Rev. Genet. 8, 639–646 (2007).
Hastings, P. J., Lupski, J. R., Rosenberg, S. M. & Ira, G. Mechanisms of change in gene copy number. Nature Rev. Genet. 10, 551–564 (2009).
Jacobs, P. A., Browne, C., Gregson, N., Joyce, C. & White, H. Estimates of the frequency of chromosome abnormalities detectable in unselected newborns using moderate levels of banding. J. Med. Genet. 29, 103–108 (1992).
Lander, E. S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
International HapMap Consortium. A haplotype map of the human genome. Nature 437, 1299–1320 (2005).
Iafrate, A. J. et al. Detection of large-scale variation in the human genome. Nature Genet. 36, 949–951 (2004).
Sebat, J. et al. Large-scale copy number polymorphism in the human genome. Science 305, 525–528 (2004). References 7 and 8 were the first to describe the genome-wide presence of CNVs in the human genome, which provided the basis for the generation of a CNV map. Reference 7 also shows that segmental duplications can be copy number variable and introduces the idea of a public CNV database.
Redon, R. et al. Global variation in copy number in the human genome. Nature 444, 444–454 (2006). This is the first comprehensive study to investigate CNV (larger than 50 kb in size) in multiple samples in order to study its impact on population genetics and genome dynamics.
Tuzun, E. et al. Fine-scale structural variation of the human genome. Nature Genet. 37, 727–732 (2005).
Levy, S. et al. The diploid genome sequence of an individual human. PLoS Biol. 5, e254 (2007). This paper reports the first personal genome sequence of an identified individual that was generated using Sanger sequencing and identifies hundreds of thousands of smaller indels in human DNA.
MacDonald, J. R., Ziman, R., Yuen, R. K., Feuk, L. & Scherer, S. W. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 42, D986–D992 (2014).
Conrad, D. F. & Hurles, M. E. The population genetics of structural variation. Nature Genet. 39, S30–S36 (2007).
Conrad, D. F. et al. Origins and functional impact of copy number variation in the human genome. Nature 464, 704–712 (2010). This paper provides a second-generation CNV (larger than 450 bp in size) map that was constructed using high-resolution oligonucleotide microarrays, which represents a 'gold-standard' data set for comparisons.
Lee, C. & Scherer, S. W. The clinical context of copy number variation in the human genome. Expert Rev. Mol. Med. 12, e8 (2010).
Pang, A. W., Macdonald, J. R., Yuen, R. K., Hayes, V. M. & Scherer, S. W. Performance of high-throughput sequencing for the discovery of genetic variation across the complete size spectrum. G3 (Bethesda) 4, 63–65 (2014).
Lupski, J. R. Genomic rearrangements and sporadic disease. Nature Genet. 39, S43–S47 (2007).
Hurles, M. E., Dermitzakis, E. T. & Tyler-Smith, C. The functional impact of structural variation in humans. Trends Genet. 24, 238–245 (2008).
Buchanan, J. A. & Scherer, S. W. Contemplating effects of genomic structural variation. Genet. Med. 10, 639–647 (2008).
Perry, G. H. et al. Diet and the evolution of human amylase gene copy number variation. Nature Genet. 39, 1256–1260 (2007).
Pinto, D. et al. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am. J. Hum. Genet. 94, 677–694 (2014).
Pinto, D. et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature 466, 368–372 (2010).
Malhotra, D. & Sebat, J. CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell 148, 1223–1241 (2012).
Wellcome Trust Case Control Consortium et al. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature 464, 713–720 (2010).
Cantsilieris, S. & White, S. J. Correlating multiallelic copy number polymorphisms with disease susceptibility. Hum. Mutat. 34, 1–13 (2013).
Jacquemont, S. et al. Mirror extreme BMI phenotypes associated with gene dosage at the chromosome 16p11.2 locus. Nature 478, 97–102 (2011).
Firth, H. V. et al. DECIPHER: database of chromosomal imbalance and phenotype in humans using ensembl resources. Am. J. Hum. Genet. 84, 524–533 (2009).
Riggs, E. R. et al. Towards an evidence-based process for the clinical interpretation of copy number variation. Clin. Genet. 81, 403–412 (2012). This paper provides an evidence-based framework for clinical evaluation, which supports or refutes the dosage sensitivity for individual genes and regions.
de Vries, B. B. et al. Diagnostic genome profiling in mental retardation. Am. J. Hum. Genet. 77, 606–616 (2005).
Lupski, J. R. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 14, 417–422 (1998).
Nuttle, X., Itsara, A., Shendure, J. & Eichler, E. E. Resolving genomic disorder-associated breakpoints within segmental DNA duplications using massively parallel sequencing. Nature Protoc. 9, 1496–1513 (2014).
Lee, C., Iafrate, A. J. & Brothman, A. R. Copy number variations and clinical cytogenetic diagnosis of constitutional disorders. Nature Genet. 39, S48–S54 (2007).
Choy, K. W., Setlur, S. R., Lee, C. & Lau, T. K. The impact of human copy number variation on a new era of genetic testing. BJOG 117, 391–398 (2010).
Miller, D. T. et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am. J. Hum. Genet. 86, 749–764 (2010).
de Leeuw, N. et al. Diagnostic interpretation of array data using public databases and internet sources. Hum. Mutat. 33, 930–940 (2012).
Church, D. M. et al. Public data archives for genomic structural variation. Nature Genet. 42, 813–814 (2010).
Campbell, I. M. et al. Parental somatic mosaicism is underrecognized and influences recurrence risk of genomic disorders. Am. J. Hum. Genet. 95, 173–182 (2014).
Forsberg, L. A. et al. Age-related somatic structural changes in the nuclear genome of human blood cells. Am. J. Hum. Genet. 90, 217–228 (2012).
Scherer, S. W. et al. Challenges and standards in integrating surveys of structural variation. Nature Genet. 39, S7–S15 (2007). This paper highlights the challenges in the characterization and documentation of structural variation. The authors propose recommendations that can be adopted for standardizing the presentation of CNVs and structural variations.
Pinto, D. et al. Comprehensive assessment of array-based platforms and calling algorithms for detection of copy number variants. Nature Biotech. 29, 512–520 (2011).
Park, H. et al. Discovery of common Asian copy number variants using integrated high-resolution array CGH and massively parallel DNA sequencing. Nature Genet. 42, 400–405 (2010).
Kidd, J. M. et al. Mapping and sequencing of structural variation from eight human genomes. Nature 453, 56–64 (2008).
Pang, A. W. et al. Towards a comprehensive structural variation map of an individual human genome. Genome Biol. 11, R52 (2010).
1000 Genomes Project Consortium et al. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012). Small deletions in the human genome have been identified in a large sample collection by the 1000 Genomes Project.
Campbell, C. D. et al. Population-genetic properties of differentiated human copy-number polymorphisms. Am. J. Hum. Genet. 88, 317–332 (2011).
Alkan, C. et al. Personalized copy number and segmental duplication maps using next-generation sequencing. Nature Genet. 41, 1061–1067 (2009).
Sudmant, P. H. et al. Diversity of human copy number variation and multicopy genes. Science 330, 641–646 (2010).
Hollox, E. J. et al. Psoriasis is associated with increased β-defensin genomic copy number. Nature Genet. 40, 23–25 (2008).
de Smith, A. J. et al. Array CGH analysis of copy number variation identifies 1284 new genes variant in healthy white males: implications for association studies of complex diseases. Hum. Mol. Genet. 16, 2783–2794 (2007).
1000 Genomes Project Consortium et al. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010).
Mills, R. E. et al. Mapping copy number variation by population-scale genome sequencing. Nature 470, 59–65 (2011).
Itsara, A. et al. Population analysis of large copy number variants and hotspots of human genetic disease. Am. J. Hum. Genet. 84, 148–161 (2009).
Ye, Y. N., Hua, Z. G., Huang, J., Rao, N. & Guo, F. B. CEG: a database of essential gene clusters. BMC Genomics 14, 769 (2013).
McCarroll, S. A. et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nature Genet. 40, 1166–1174 (2008).
Stankiewicz, P. & Lupski, J. R. Structural variation in the human genome and its role in disease. Annu. Rev. Med. 61, 437–455 (2010).
Makino, T., McLysaght, A. & Kawata, M. Genome-wide deserts for copy number variation in vertebrates. Nature Commun. 4, 2283 (2013).
Yuen, R. K. et al. Development of a high-resolution Y-chromosome microarray for improved male infertility diagnosis. Fertil. Steril. 101, 1079–1085. e3 (2014).
Wong, L. P. et al. Deep whole-genome sequencing of 100 southeast Asian Malays. Am. J. Hum. Genet. 92, 52–66 (2013).
ENCODE Project Consortium et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816 (2007).
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Gerstein, M. B. et al. Comparative analysis of the transcriptome across distant species. Nature 512, 445–448 (2014).
Pruitt, K. D., Tatusova, T. & Maglott, D. R. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 35, D61–D65 (2007).
Johansson, A. C. & Feuk, L. Characterization of copy number-stable regions in the human genome. Hum. Mutat. 32, 947–955 (2011). This paper defines a list of dosage-sensitive regions of the human genome and correlates them with the rare and de novo CNVs identified in patients with intellectual disability or autism.
Petrovski, S., Wang, Q., Heinzen, E. L., Allen, A. S. & Goldstein, D. B. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 9, e1003709 (2013).
Huang, N., Lee, I., Marcotte, E. M. & Hurles, M. E. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 6, e1001154 (2010). This important study uses functional features to determine haploinsufficiency scores for human protein-coding genes and their likelihood to be involved in disease.
Hurles, M. Gene duplication: the genomic trade in spare parts. PLoS Biol. 2, E206 (2004).
Nguyen, D. Q., Webber, C. & Ponting, C. P. Bias of selection on human copy-number variants. PLoS Genet. 2, e20 (2006).
Ng, P. C. et al. Genetic variation in an individual human exome. PLoS Genet. 4, e1000160 (2008).
Katzman, S. et al. Human genome ultraconserved elements are ultraselected. Science 317, 915 (2007).
Nguyen, D. Q. et al. Reduced purifying selection prevails over positive selection in human copy number variant evolution. Genome Res. 18, 1711–1723 (2008).
Uddin, M. et al. Brain-expressed exons under purifying selection are enriched for de novo mutations in autism spectrum disorder. Nature Genet. 46, 742–747 (2014).
Bailey, J. A. et al. Recent segmental duplications in the human genome. Science 297, 1003–1007 (2002). This paper provides the first map of segmental duplications in the human genome, which includes an analysis of their relationship to genes and genetic diseases.
Mefford, H. C. & Eichler, E. E. Duplication hotspots, rare genomic disorders, and common disease. Curr. Opin. Genet. Dev. 19, 196–204 (2009).
Woods, S. et al. Duplication and retention biases of essential and non-essential genes revealed by systematic knockdown analyses. PLoS Genet. 9, e1003330 (2013).
Yang, T. L. et al. Genome-wide copy-number-variation study identified a susceptibility gene, UGT2B17, for osteoporosis. Am. J. Hum. Genet. 83, 663–674 (2008).
Szulman, A., Nardozza, L. M., Barreto, J. A., Araujo Junior, E. & Moron, A. F. Investigation of pseudogenes RHDΨ and RHD-CE-D hybrid gene in D-negative blood donors by the real time PCR method. Transfus Apher Sci. 47, 289–293 (2012).
Jiang, Y. et al. KIR3DS1/L1 and HLA-Bw4-80I are associated with HIV disease progression among HIV typical progressors and long-term nonprogressors. BMC Infect. Dis. 13, 405 (2013).
International Multiple Sclerosis Genetics Consortium et al. Risk alleles for multiple sclerosis identified by a genomewide study. N. Engl. J. Med. 357, 851–862 (2007).
Hadithi, M. et al. Accuracy of serologic tests and HLA-DQ typing for diagnosing celiac disease. Ann. Intern. Med. 147, 294–302 (2007).
Bartels, I. & Lindemann, A. Maternal levels of pregnancy-specific β1-glycoprotein (SP-1) are elevated in pregnancies affected by Down's syndrome. Hum. Genet. 80, 46–48 (1988).
MacArthur, D. G. et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science 335, 823–828 (2012).
Ashburner, M. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet. 25, 25–29 (2000).
Aguiar, D., Halldorsson, B. V., Morrow, E. M. & Istrail, S. DELISHUS: an efficient and exact algorithm for genome-wide detection of deletion polymorphism in autism. Bioinformatics 28, i154–i162 (2012).
Cooper, G. M., Nickerson, D. A. & Eichler, E. E. Mutational and selective effects on copy-number variants in the human genome. Nature Genet. 39, S22–S29 (2007).
McKernan, K. J. et al. Sequence and structural variation in a human genome uncovered by short-read, massively parallel ligation sequencing using two-base encoding. Genome Res. 19, 1527–1541 (2009).
Iskow, R. C., Gokcumen, O. & Lee, C. Exploring the role of copy number variants in human adaptation. Trends Genet. 28, 245–257 (2012).
Hasin-Brumshtein, Y., Lancet, D. & Olender, T. Human olfaction: from genomic variation to phenotypic diversity. Trends Genet. 25, 178–184 (2009).
Blekhman, R. et al. Natural selection on genes that underlie human disease susceptibility. Curr. Biol. 18, 883–889 (2008).
International HapMap 3 Consortium et al. Integrating common and rare genetic variation in diverse human populations. Nature 467, 52–58 (2010).
Nelson, M. R. et al. An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people. Science 337, 100–104 (2012).
Lionel, A. C. et al. Disruption of the ASTN2/TRIM32 locus at 9q33.1 is a risk factor in males for autism spectrum disorders, ADHD and other neurodevelopmental phenotypes. Hum. Mol. Genet. 23, 2752–2768 (2014).
Kuningas, M. et al. Large common deletions associate with mortality at old age. Hum. Mol. Genet. 20, 4290–4296 (2011).
Costain, G. et al. Adult neuropsychiatric expression and familial segregation of 2q13 duplications. Am. J. Med. Genet. B Neuropsychiatr. Genet. 165B, 337–344 (2014).
Castoldi, G. L., Ricci, N., Punturieri, E. & Bosi, L. Chromosomal imbalance in plasmacytoma. Lancet 1, 829 (1963).
Cattanach, B. M. Snaker: a dominant abnormality caused by chromosomal imbalance. Z. Vererbungsl. 96, 275–284 (1965).
Sparkes, R. S. Genetic abnormalities: the consequences of chromosome imbalance. Science 235, 916a (1987).
Epstein, C. J. The consequences of chromosome imbalance. Am. J. Med. Genet. Suppl. 7, 31–37 (1990).
Bejjani, B. A., Theisen, A. P., Ballif, B. C. & Shaffer, L. G. Array-based comparative genomic hybridization in clinical diagnosis. Expert Rev. Mol. Diagn. 5, 421–429 (2005).
Merico, D., Isserlin, R., Stueker, O., Emili, A. & Bader, G. D. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS ONE 5, e13984 (2010).
Acknowledgements
The authors thank R. Ziman and G. Pellecchia for computational support, as well as J. Buchanan, J. Stavropoulos, C. Marshall, R. Yuen, B. Thiruvahindrapuram, M. Uddin, M. Mohammed and L. Feuk for discussions. They thank The Centre for Applied Genomics Science and Technology Innovation Centre (funded by Genome Canada and the Ontario Genomics Institute) for computational support. The Database of Genomic Variants and our research are supported by grants from Genome Canada, the Canada Foundation of Innovation, the Canadian Institute for Advanced Research, the government of Ontario, the Canadian Institutes of Health Research (CIHR), The Hospital for Sick Children, and the University of Toronto McLaughlin Centre. S.W.S. holds the GlaxoSmithKline–CIHR Endowed Chair in Genome Sciences at The Hospital for Sick Children and the University of Toronto.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary Text and Figures (PDF 5443 kb)
Supplementary information Table S1
(XLS 55 kb)
Supplementary information Table S3
(XLS 55 kb)
Supplementary information Table S4
(TXT 2847 kb)
Supplementary information Table S5
(XLS 454 kb)
Supplementary information Table S6
(XLSX 2474 kb)
Supplementary information Table S7
(XLSX 864 kb)
Supplementary information Table S8
(XLSX 2352 kb)
Supplementary information Table S9
(XLS 7414 kb)
Supplementary information Table S10
(XLS 3592 kb)
Supplementary information Table S11
(XLS 52 kb)
Supplementary information Table S13
(XLS 355 kb)
Supplementary information Table S15
(XLS 1566 kb)
Supplementary information Table S17
(XLS 137 kb)
Supplementary information Table S18
(XLS 135 kb)
Supplementary information Table S19
(XLS 4666 kb)
Glossary
- Copy number variation
-
(CNV). A genomic segment of at least 50 bp that differs in copy number based on the comparison of two or more genomes.
- Unbalanced rearrangements
-
Genomic variants that involve loss (deletion) or gain (duplication) of segments of the genome.
- Database of Genomic Variants
-
(DGV). A curated catalogue of copy number and structural variations in the human genomes of healthy control individuals.
- Copy number variable regions
-
(CNVRs). Regions containing at least two copy number variations that overlap and that may have different breakpoints.
- Next-generation sequencing
-
(NGS). A high-throughput DNA sequencing technology that typically generates shorter reads than Sanger sequencing-based methods and that can sequence billions of bases in parallel. NGS minimizes the need for fragment cloning.
- Comparative genomic hybridization
-
(CGH). An array-based technique that interrogates the genome for signs of deletion or duplication in relation to a reference.
- SNP-based arrays
-
Single-nucleotide polymorphism (SNP)-based microarrays that contain SNP probes to genotype human DNA at the single-base level. However, through dosage signals in adjacent regions, they can be used to recognize copy number variations.
- Segmental duplications
-
(Also known as low-copy repeats). Highly homologous duplicated segments of DNA that are >1 kb in length and that show >90% sequence similarity.
- International Standards for Cytogenomic Arrays
-
(ISCA). A consortium of clinical cytogeneticists who work together to standardize the use of array-based approaches in clinical genetic testing.
- Database of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources
-
(DECIPHER). A database that documents phenotype information in patients with observed chromosome abnormalities and that aids the interpretation of genomic variants.
- DECIPHER critical genes
-
Genes located in the critical regions that are associated with the 70 syndromes defined in Database of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources (DECIPHER).
- Essential genes
-
Orthologues of mouse genes for which homozygous loss-of-function mutations cause embryonic or neonatal lethality. They are necessary for cellular viability and organism development. They are evolutionarily more conserved than non-essential genes.
- Copy number stable
-
(CNS). Pertaining to regions of the genome without any detected copy number variation in healthy individuals.
- Genic intolerance score
-
An index of intolerance to rare, non-synonymous variation.
- Haploinsufficiency
-
Reduction in the amount of gene product owing to functional loss of an allele that leads to an abnormal or a disease state.
- Long intergenic non-coding RNAs
-
(lincRNAs). Non-coding RNAs that are thought to be key regulators of diverse cellular processes. Their expression seems to be more tissue-specific than that of coding genes.
- PhastCons elements
-
Evolutionarily conserved elements that were identified by modelling substitution rates in multiple genome alignments.
- Ultra-conserved elements
-
Regions of DNA that are conserved across mammalian genomes and that mostly consist of non-protein-coding regions (that is, regions with little or no evolutionary changes since the divergence of mammals and birds).
Rights and permissions
About this article
Cite this article
Zarrei, M., MacDonald, J., Merico, D. et al. A copy number variation map of the human genome. Nat Rev Genet 16, 172–183 (2015). https://doi.org/10.1038/nrg3871
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg3871