Key Points
-
Each tumour represents an independent evolutionary experiment, starting from almost the same point: namely, the fertilized egg. As tools for genome-wide analysis become more widely applied, commonalities and variations in the patterns of cancer evolution are emerging, and this has important implications for our understanding of cancer biology and the management of patients with tumours.
-
Evolution may be investigated within an individual cancer through various sampling approaches, ranging from a single sample to multiple samples taken over space and/or time or even from multiple single cells.
-
Massively parallel sequencing data from bulk tumour sampling lend themselves to mathematical approaches that permit the reconstruction of the underlying genomic architecture.
-
Genomic technologies have identified extreme genomic heterogeneity between and even within tumour types. This suggests that evolutionary pathways underlying cancers are diverse and highlights one of the challenges for the design of cancer therapies.
-
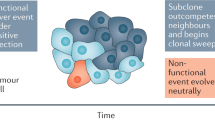
Intratumoural heterogeneity reflects the branching and dynamic nature of cancer evolution.
-
When a new mutation arises in a cancer cell, the subsequent evolutionary trajectory of the cell will be influenced by the cellular ground state and any pre-existing mutations. Understanding epistatic interactions that operate within a cancer cell will contribute to our comprehension of carcinogenesis and is important for designing targeted therapeutic approaches.
-
Emerging observations suggest that cancers do not necessarily arise gradually through multiple steps but that sudden 'crisis' events can accelerate carcinogenesis.
-
Many of the aggressive clinical characteristics of cancer depend on the continued generation of variation. There is evidence for the existence of genomic instability in many cancer types, but it remains unclear whether it is a pre-requisite for cancer development or whether cancers can evolve in the presence of a normal mutation rate.
Abstract
The advent of massively parallel sequencing technologies has allowed the characterization of cancer genomes at an unprecedented resolution. Investigation of the mutational landscape of tumours is providing new insights into cancer genome evolution, laying bare the interplay of somatic mutation, adaptation of clones to their environment and natural selection. These studies have demonstrated the extent of the heterogeneity of cancer genomes, have allowed inferences to be made about the forces that act on nascent cancer clones as they evolve and have shown insight into the mutational processes that generate genetic variation. Here we review our emerging understanding of the dynamic evolution of the cancer genome and of the implications for basic cancer biology and the development of antitumour therapy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Nowell, P. C. The clonal evolution of tumor cell populations. Science 194, 23–28 (1976).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Vogelstein, B. & Kinzler, K. W. The multistep nature of cancer. Trends Genet. 9, 138–141 (1993).
Stratton, M. R., Campbell, P. J. & Futreal, P. A. The cancer genome. Nature 458, 719–724 (2009).
Campbell, P. J. et al. Subclonal phylogenetic structures in cancer revealed by ultra-deep sequencing. Proc. Natl Acad. Sci. USA 105, 13081–13086 (2008).
Pleasance, E. D. et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature 463, 191–196 (2010).
Ding, L. et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 464, 999–1005 (2010).
Turajlic, S. et al. Whole genome sequencing of matched primary and metastatic acral melanomas. Genome Res. 22, 196–207 (2012).
Gerlinger, M. et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366, 883–892 (2012).
Navin, N. et al. Tumour evolution inferred by single-cell sequencing. Nature 472, 90–94 (2011).
Yachida, S. et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 467, 1114–1117 (2010).
Campbell, P. J. et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature 467, 1109–1113 (2010).
Hou, Y. et al. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell 148, 873–885 (2012).
Xu, X. et al. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell 148, 886–895 (2012). References 13 and 14 provide proof of principle that next-generation sequencing technologies can be combined with single-cell approaches can be used to investigate intra-tumoural heterogeneity.
Anderson, K. et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature 469, 356–361 (2011).
Baker, M. Digital PCR hits its stride. Nature Methods 9, 541–544 (2012).
Wang, J. et al. Quantifying EGFR alterations in the lung cancer genome with nanofluidic digital PCR arrays. Clin. Chem. 56, 623–632 (2010).
Anderson, A. R., Weaver, A. M., Cummings, P. T. & Quaranta, V. Tumor morphology and phenotypic evolution driven by selective pressure from the microenvironment. Cell 127, 905–915 (2006). This is a comprehensive review of the literature from the field of mathematical modelling in cancer evolution.
Michor, F., Iwasa, Y. & Nowak, M. A. Dynamics of cancer progression. Nature Rev. Cancer 4, 197–205 (2004).
Attolini, C. S. & Michor, F. Evolutionary theory of cancer. Ann. NY Acad. Sci. 1168, 23–51 (2009).
Ding, L. et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 481, 506–510 (2012).
Carter, S. L. et al. Absolute quantification of somatic DNA alterations in human cancer. Nature Biotech. 30, 413–421 (2012).
Durinck, S. et al. Temporal dissection of tumorigenesis in primary cancers. Cancer Discov. 1, 137–143 (2011).
Nik-Zainal, S. et al. The life history of 21 breast cancers. Cell 149, 994–1007 (2012).
Nik-Zainal, S. et al. Mutational processes molding the genomes of 21 breast cancers. Cell 149, 979–993 (2012).
Greenman, C. D. et al. Estimation of rearrangement phylogeny for cancer genomes. Genome Res. 22, 346–361 (2012). In this article, a mathematical framework is presented for reconstructing temporal sequences of rearrangements and hence evolutionary selection.
Futreal, P. A. et al. A census of human cancer genes. Nature Rev. Cancer 4, 177–183 (2004).
Blanquet, V. et al. Spectrum of germline mutations in the RB1 gene: a study of 232 patients with hereditary and non hereditary retinoblastoma. Hum. Mol. Genet. 4, 383–388 (1995).
Persson, M. et al. Recurrent fusion of MYB and NFIB transcription factor genes in carcinomas of the breast and head and neck. Proc. Natl Acad. Sci. USA 106, 18740–18744 (2009).
Ellis, M. J. et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature 486, 353–360 (2012). This was one of the first studies to correlate somatic mutation changes identified through next-generation sequencing, with treatment responses. Somatic mutations are also mapped to distinct pathways of relevance to tumour cell biology.
Curtis, C. et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352 (2012). This study is an integrated analysis of copy number and gene expression data with long-term clinical follow-up providing a novel molecular stratification of the breast cancer population.
Shah, S. P. et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature 486, 395–399 (2012).
Perreard, L. et al. Classification and risk stratification of invasive breast carcinomas using a real-time quantitative RT-PCR assay. Breast Cancer Res. 8, R23 (2006).
Parsons, D. W. et al. The genetic landscape of the childhood cancer medulloblastoma. Science 331, 435–439 (2011).
Zhang, J. et al. A novel retinoblastoma therapy from genomic and epigenetic analyses. Nature 481, 329–334 (2012).
Ley, T. J. et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature 456, 66–72 (2008).
Mardis, E. R. et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N. Engl. J. Med. 361, 1058–1066 (2009).
Ng, C. K. et al. The role of tandem duplicator phenotype in tumour evolution in high-grade serous ovarian cancer. J. Pathol. 226, 703–712 (2012).
McBride, D. J. et al. Tandem duplication of chromosomal segments is common in ovarian and breast cancer genomes. J. Pathol. 227, 446–455 (2012).
Berger, M. F. et al. The genomic complexity of primary human prostate cancer. Nature 470, 214–220 (2011).
Stephens, P. J. et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144, 27–40 (2011).
Rausch, T. et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell 148, 59–71 (2012).
Molenaar, J. J. et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature 483, 589–593 (2012).
Lengauer, C., Kinzler, K. W. & Vogelstein, B. Genetic instabilities in human cancers. Nature 396, 643–649 (1998).
Pleasance, E. D. et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature 463, 184–190 (2010).
Gatenby, R. A. et al. Cellular adaptations to hypoxia and acidosis during somatic evolution of breast cancer. Br. J. Cancer 97, 646–653 (2007).
Schwartz, J. L., Jordan, R., Sun, J., Ma, H. & Hsieb, A. W. Dose-dependent changes in the spectrum of mutations induced by ionizing radiation. Radiat. Res. 153, 312–317 (2000).
DeMarini, D. M. Genotoxicity of tobacco smoke and tobacco smoke condensate: a review. Mutat. Res. 567, 447–474 (2004).
Le Calvez, F. et al. TP53 and KRAS mutation load and types in lung cancers in relation to tobacco smoke: distinct patterns in never, former, and current smokers. Cancer Res. 65, 5076–5083 (2005).
Arkenau, H. T., Kefford, R. & Long, G. V. Targeting BRAF for patients with melanoma. Br. J. Cancer 104, 392–398 (2011).
Su, X. et al. Cascading adoptive cell therapy for metastatic melanoma. Cancer Biother. Radiopharm. 26, 401–406 (2011).
Coufal, N. G. et al. L1 retrotransposition in human neural progenitor cells. Nature 460, 1127–1131 (2009).
Srinivasan, D. & Plattner, R. Activation of ABL tyrosine kinases promotes invasion of aggressive breast cancer cells. Cancer Res. 66, 5648–5655 (2006).
Antonescu, C. R. Gastrointestinal stromal tumor (GIST) pathogenesis, familial GIST, and animal models. Semin. Diagn. Pathol. 23, 63–69 (2006).
Kwon, J. G. et al. Changes in the structure and function of ICC networks in ICC hyperplasia and gastrointestinal stromal tumors. Gastroenterology 136, 630–639 (2009).
Chi, P. et al. ETV1 is a lineage survival factor that cooperates with KIT in gastrointestinal stromal tumours. Nature 467, 849–853 (2010).
Banerji, S. et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature 486, 405–409 (2012).
Greenblatt, M. S., Chappuis, P. O., Bond, J. P., Hamel, N. & Foulkes, W. D. TP53 mutations in breast cancer associated with BRCA1 or BRCA2 germ-line mutations: distinctive spectrum and structural distribution. Cancer Res. 61, 4092–4097 (2001).
Matsumoto, S. et al. Prevalence and specificity of LKB1 genetic alterations in lung cancers. Oncogene 26, 5911–5918 (2007).
Mahoney, C. L. et al. LKB1/KRAS mutant lung cancers constitute a genetic subset of NSCLC with increased sensitivity to MAPK and mTOR signalling inhibition. Br. J. Cancer 100, 370–375 (2009).
Jones, C. J. et al. Evidence for a telomere-independent “clock” limiting RAS oncogene-driven proliferation of human thyroid epithelial cells. Mol. Cell. Biol. 20, 5690–5699 (2000).
Serrano, M., Lin, A. W., McCurrach, M. E., Beach, D. & Lowe, S. W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88, 593–602 (1997).
Jacobs, J. J. et al. Senescence bypass screen identifies TBX2, which represses CDKN2A (p19(ARF)) and is amplified in a subset of human breast cancers. Nature Genet. 26, 291–299 (2000).
Vance, K. W., Carreira, S., Brosch, G. & Goding, C. R. Tbx2 is overexpressed and plays an important role in maintaining proliferation and suppression of senescence in melanomas. Cancer Res. 65, 2260–2268 (2005).
Notta, F. et al. Evolution of human BCR–ABL1 lymphoblastic leukaemia-initiating cells. Nature 469, 362–367 (2011).
Bridgham, J. T., Ortlund, E. A. & Thornton, J. W. An epistatic ratchet constrains the direction of glucocorticoid receptor evolution. Nature 461, 515–519 (2009).
Moore, J. H. A global view of epistasis. Nature Genet. 37, 13–14 (2005).
Bissonnette, R. P., Echeverri, F., Mahboubi, A. & Green, D. R. Apoptotic cell death induced by c-MYC is inhibited by BCL-2. Nature 359, 552–554 (1992).
Fanidi, A., Harrington, E. A. & Evan, G. I. Cooperative interaction between c-MYC and BCL-2 proto-oncogenes. Nature 359, 554–556 (1992).
Rehman, F. L., Lord, C. J. & Ashworth, A. Synthetic lethal approaches to breast cancer therapy. Nature Rev. Clin. Oncol. 7, 718–724 (2010).
Farmer, H. et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 (2005).
Swisher, E. M. et al. Secondary BRCA1 mutations in BRCA1-mutated ovarian carcinomas with platinum resistance. Cancer Res. 68, 2581–2586 (2008).
Sakai, W. et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature 451, 1116–1120 (2008).
Artandi, S. E. et al. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature 406, 641–645 (2000).
Bignell, G. R. et al. Architectures of somatic genomic rearrangement in human cancer amplicons at sequence-level resolution. Genome Res. 17, 1296–1303 (2007).
Rudolph, K. L., Millard, M., Bosenberg, M. W. & DePinho, R. A. Telomere dysfunction and evolution of intestinal carcinoma in mice and humans. Nature Genet. 28, 155–159 (2001).
Crasta, K. et al. DNA breaks and chromosome pulverization from errors in mitosis. Nature 482, 53–58 (2012).
Magrangeas, F., Avet-Loiseau, H., Munshi, N. C. & Minvielle, S. Chromothripsis identifies a rare and aggressive entity among newly diagnosed multiple myeloma patients. Blood 118, 675–678 (2011).
Beerenwinkel, N. et al. Genetic progression and the waiting time to cancer. PLoS Comput. Biol. 3, e225 (2007).
Sjoblom, T. et al. The consensus coding sequences of human breast and colorectal cancers. Science 314, 268–274 (2006).
Welch, J. S. et al. The origin and evolution of mutations in acute myeloid leukemia. Cell 150, 264–278 (2012). This study presents mutational data from normal haematopoietic stem cells, which were found to show similar mutational burden and signatures to those seen in acute leukaemias.
Martin, S. A., Hewish, M., Lord, C. J. & Ashworth, A. Genomic instability and the selection of treatments for cancer. J. Pathol. 220, 281–289 (2010).
Vilar, E. & Gruber, S. B. Microsatellite instability in colorectal cancer—the stable evidence. Nature Rev. Clin. Oncol. 7, 153–162 (2010).
Sheltzer, J. M. et al. Aneuploidy drives genomic instability in yeast. Science 333, 1026–1030 (2011).
Solomon, D. A. et al. Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science 333, 1039–1043 (2011).
Gordon, D. J., Resio, B. & Pellman, D. Causes and consequences of aneuploidy in cancer. Nature Rev. Genet. 13, 189–203 (2012).
Cheng, Y. W. et al. CpG island methylator phenotype associates with low-degree chromosomal abnormalities in colorectal cancer. Clin. Cancer Res. 14, 6005–6013 (2008).
Noushmehr, H. et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 17, 510–522 (2010).
Loeb, L. A., Bielas, J. H. & Beckman, R. A. Cancers exhibit a mutator phenotype: clinical implications. Cancer Res. 68, 3551–3557 (2008).
Bodmer, W., Bielas, J. H. & Beckman, R. A. Genetic instability is not a requirement for tumor development. Cancer Res. 68, 3558–3560 (2008).
Jones, D. T. et al. Dissecting the genomic complexity underlying medulloblastoma. Nature 488, 100–105 (2012).
Pasqualucci, L. et al. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature 412, 341–346 (2001).
Pasqualucci, L. et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nature Genet. 43, 830–837 (2011).
Migliazza, A. et al. Frequent somatic hypermutation of the 5′ noncoding region of the BCL6 gene in B-cell lymphoma. Proc. Natl Acad. Sci. USA 92, 12520–12524 (1995).
Bignell, G. R. et al. Signatures of mutation and selection in the cancer genome. Nature 463, 893–898 (2010).
Gandhi, M., Dillon, L. W., Pramanik, S., Nikiforov, Y. E. & Wang, Y. H. DNA breaks at fragile sites generate oncogenic RET/PTC rearrangements in human thyroid cells. Oncogene 29, 2272–2280 (2010).
Letessier, A. et al. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature 470, 120–123 (2011).
Lang, G. I. & Murray, A. W. Mutation rates across budding yeast chromosome VI are correlated with replication timing. Genome Biol. Evol. 3, 799–811 (2011).
Veeriah, S. et al. Somatic mutations of the Parkinson's disease-associated gene PARK2 in glioblastoma and other human malignancies. Nature Genet. 42, 77–82 (2010).
Poulogiannis, G. et al. PARK2 deletions occur frequently in sporadic colorectal cancer and accelerate adenoma development in Apc mutant mice. Proc. Natl Acad. Sci. USA 107, 15145–15150 (2010).
Neves, H., Ramos, C., da Silva, M. G., Parreira, A. & Parreira, L. The nuclear topography of ABL, BCR, PML, and RARα genes: evidence for gene proximity in specific phases of the cell cycle and stages of hematopoietic differentiation. Blood 93, 1197–1207 (1999).
Mani, R. S. et al. Induced chromosomal proximity and gene fusions in prostate cancer. Science 326, 1230 (2009).
Lin, C. et al. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell 139, 1069–1083 (2009).
Stephens, P. J. et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature 462, 1005–1010 (2009).
Markowitz, S. et al. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science 268, 1336–1338 (1995).
Chapman, M. A. et al. Initial genome sequencing and analysis of multiple myeloma. Nature 471, 467–472 (2011).
Dunson, D. B. Nonparametric Bayes Applications to Biostatistics (Cambridge Univ. Press, 2010).
Weisenberger, D. J. et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nature Genet. 38, 787–793 (2006).
Flanagan, J. M. et al. Intra- and interindividual epigenetic variation in human germ cells. Am. J. Hum. Genet. 79, 67–84 (2006).
Ji, H. et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature 467, 338–342 (2010).
Jones, S. et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science 330, 228–231 (2010).
Wiegand, K. C. et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N. Engl. J. Med. 363, 1532–1543 (2010).
Dalgliesh, G. L. et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature 463, 360–363 (2010).
Morin, R. D. et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nature Genet. 42, 181–185 (2010).
Ernst, T. et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nature Genet. 42, 722–726 (2010).
Figueroa, M. E. et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell 17, 13–27 (2010).
Acknowledgements
We would like to thank the Wellcome Trust for funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- Mutational signatures
-
Patterns of mutations that are characteristic of a type of cancer or that are indicative of a specific process.
- Chromothripsis
-
A single event that causes genome shattering and reassembly, resulting in a characteristic pattern of oscillating copy number and up to several hundred genomic rearrangements localized to one or a few chromosomes.
- Driver mutations
-
Somatic mutations within cancer genes that confer a clonal advantage, that are causally implicated in oncogenesis and that are positively selected for during cancer evolution.
- Synthetic lethality
-
Two genes are synthetically lethal if mutation of either in isolation is compatible with viability, but mutation of both leads to cell death.
- Kataegis
-
A localized hypermutation that often colocalizes with somatic rearrangements.
- Microsatellite instability
-
(MSI). Microsatellites are repeating sequences within DNA of 2–6 base pairs in length; defects in mismatch repair can give rise to genomic instability within these regions.
- Chromosomal instability
-
(CIN). A form of genomic instability that is common in cancers and is characterized by large chromosomal losses by as of yet undefined mechanisms.
Rights and permissions
About this article
Cite this article
Yates, L., Campbell, P. Evolution of the cancer genome. Nat Rev Genet 13, 795–806 (2012). https://doi.org/10.1038/nrg3317
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg3317
This article is cited by
-
Label-free quantification of imaging features in the extracellular matrix of left and right-sided colon cancer tissues
Scientific Reports (2024)
-
Impact of Resistance on Therapeutic Design: A Moran Model of Cancer Growth
Bulletin of Mathematical Biology (2024)
-
Rapid assessment of 3-dimensional intra-tumor heterogeneity through cycling temperature capillary electrophoresis
BMC Research Notes (2023)
-
Evolution of intra-tumoral heterogeneity across different pathological stages in papillary thyroid carcinoma
Cancer Cell International (2022)
-
Uphyloplot2: visualizing phylogenetic trees from single-cell RNA-seq data
BMC Genomics (2021)