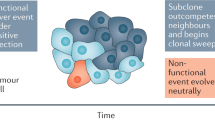
Abstract
Clonal populations of mammalian cells are inherently heterogeneous. They contain cells that display non-genetic variability resulting from gene expression noise and the fact that gene networks have multiple stable states. These stable, heritable variants within one cell type can exhibit different levels of responsiveness to environmental conditions. Hence, they could in principle serve as a temporary substrate for natural selection in the absence of mutations. We suggest that such ubiquitous but non-genetic variability can contribute to the somatic evolution of cancer cells, hence accelerating tumour progression independently of genetic mutations.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Curtis, H. J. Formal discussion of: somatic mutations and carcinogenesis. Cancer Res. 25, 1305–1308 (1965).
Cairns, J. Mutation selection and the natural history of cancer. Nature 255, 197–200 (1975).
Bergers, G., Song, S., Meyer-Morse, N., Bergsland, E. & Hanahan, D. Benefits of targeting both pericytes and endothelial cells in the tumor vasculature with kinase inhibitors. J. Clin. Invest. 111, 1287–1295 (2003).
Soto, A. M. & Sonnenschein, C. The somatic mutation theory of cancer: growing problems with the paradigm? Bioessays 26, 1097–1107 (2004).
Bernards, R. & Weinberg, R. A progression puzzle. Nature 418, 823 (2002).
Weigelt, B. & van't Veer, L. J. Hard-wired genotype in metastatic breast cancer. Cell Cycle 3, 756–757 (2004).
Liotta, L. A. & Kohn, E. C. Cancer's deadly signature. Nature Genet. 33, 10–11 (2003).
Podsypanina, K. et al. Seeding and propagation of untransformed mouse mammary cells in the lung. Science 321, 1841–1844 (2008).
Huang, S. & Ingber, D. E. A non-genetic basis for cancer progression and metastasis: self-organizing attractors in cell regulatory networks. Breast Dis. 26, 27–54 (2006).
Hanahan, D. & Weinberg, R. A. The hallmarks of cancer. Cell 100, 57–70 (2000).
Chang, H. H., Hemberg, M., Barahona, M., Ingber, D. E. & Huang, S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature 453, 544–547 (2008).
Kaern, M., Elston, T. C., Blake, W. J. & Collins, J. J. Stochasticity in gene expression: from theories to phenotypes. Nature Rev. Genet. 6, 451–464 (2005).
Patrascioiu, A. The ergodic-hypothesis, a complicated problem in mathematics and physics. Los Alamos Science 15, 263–279 (1987).
Elowitz, M. B., Levine, A. J., Siggia, E. D. & Swain, P. S. Stochastic gene expression in a single cell. Science 297, 1183–1186 (2002).
Rosenfeld, N., Young, J. W., Alon, U., Swain, P. S. & Elowitz, M. B. Gene regulation at the single-cell level. Science 307, 1962–1965 (2005).
Yu, J., Xiao, J., Ren, X., Lao, K. & Xie, X. S. Probing gene expression in live cells, one protein molecule at a time. Science 311, 1600–1603 (2006).
Blake, W. J., KÆrn, M., Cantor, C. R. & Collins, J. J. Noise in eukaryotic gene expression. Nature 422, 633–637 (2003).
Golding, I., Paulsson, J., Zawilski, S. M. & Cox, E. C. Real-time kinetics of gene activity in individual bacteria. Cell 123, 1025–1036 (2005).
Austin, D. W. et al. Gene network shaping of inherent noise spectra. Nature 439, 608–611 (2006).
Takasuka, N., White, M. R., Wood, C. D., Robertson, W. R. & Davis, J. R. Dynamic changes in prolactin promoter activation in individual living lactotrophic cells. Endocrinology 139, 1361–1368 (1998).
Dietrich, J. E. & Hiiragi, T. Stochastic patterning in the mouse pre-implantation embryo. Development 134, 4219–4231 (2007).
Sigal, A. et al. Variability and memory of protein levels in human cells. Nature 444, 643–646 (2006).
Huang, S. & Wikswo, J. Dimensions of systems biology. Rev. Physiol. Biochem. Pharmacol. 157, 81–104 (2006).
Huang, S. & Kauffman, S. in Encyclopedia of Complexity and Systems Science (ed. Meyers, R. A.) (Springer) (in the press).
Waddington, C. H. The Strategy of the Genes (Allen and Unwin, London, 1957).
Kauffman, S. Homeostasis and differentiation in random genetic control networks. Nature 224, 177–178 (1969).
Huang, S., Eichler, G., Bar-Yam, Y. & Ingber, D. E. Cell fates as high-dimensional attractor states of a complex gene regulatory network. Phys. Rev. Lett. 94, 128701 (2005).
Achilles, E. G. et al. Heterogeneity of angiogenic activity in a human liposarcoma: a proposed mechanism for “no take” of human tumors in mice. J. Natl. Cancer Inst. 93, 1075–1081 (2001).
Hill, R. P., Chambers, A. F., Ling, V. & Harris, J. F. Dynamic heterogeneity: rapid generation of metastatic variants in mouse B16 melanoma cells. Science 224, 998–1001 (1984).
Minn, A. J. et al. Distinct organ-specific metastatic potential of individual breast cancer cells and primary tumors. J. Clin. Invest. 115, 44–55 (2005).
Axelrod, D. E., Majumdar, S. K., Wivell, J. A. & Terry, R. W. Tumorigenicity of Friend murine erythroleukemia cell lines differing in spontaneous differentiation rates. Int. J. Cancer 26, 799–804 (1980).
Chow, D. A. & Greenberg, A. H. The generation of tumor heterogeneity in vivo. Int. J. Cancer 25, 261–265 (1980).
Lengauer, C., Kinzler, K. W. & Vogelstein, B. Genetic instabilities in human cancers. Nature 396, 643–649 (1998).
Dick, J. E. Stem cell concepts renew cancer research. Blood 112, 4793–4807 (2008).
Bonnet, D. & Dick, J. E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nature Med. 3, 730–737 (1997).
Campbell, L. L. & Polyak, K. Breast tumor heterogeneity: cancer stem cells or clonal evolution? Cell Cycle 6, 2332–2338 (2007).
Kauffman, S. Differentiation of malignant to benign cells. J. Theor. Biol. 31, 429–451 (1971).
Enver, T., Heyworth, C. M. & Dexter, T. M. Do stem cells play dice? Blood 92, 348–351 (1998); discussion 352.
Hume, D. A. Probability in transcriptional regulation and its implications for leukocyte differentiation and inducible gene expression. Blood 96, 2323–2328 (2000).
Dean, A. C. & Hinshelwood, C. The stability of various adaptations of Bact. lactis aerogenes (Aerobacter aerogenes). Proc. R. Soc. Lond. B 142, 45–60 (1954).
Spudich, J. L. & Koshland, D. E. Jr. Non-genetic individuality: chance in the single cell. Nature 262, 467–471 (1976).
Balaban, N. Q., Merrin, J., Chait, R., Kowalik, L. & Leibler, S. Bacterial persistence as a phenotypic switch. Science 305, 1622–1625 (2004).
Blake, W. J. et al. Phenotypic consequences of promoter-mediated transcriptional noise. Mol. Cell 24, 853–865 (2006).
Smith, M. C., Sumner, E. R. & Avery, S. V. Glutathione and Gts1p drive beneficial variability in the cadmium resistances of individual yeast cells. Mol. Microbiol. 66, 699–712 (2007).
Li, Y. & Zhang, Y. PhoU is a persistence switch involved in persister formation and tolerance to multiple antibiotics and stresses in Escherichia coli. Antimicrob. Agents Chemother. 51, 2092–2099 (2007).
Kashiwagi, A., Urabe, I., Kaneko, K. & Yomo, T. Adaptive response of a gene network to environmental changes by fitness-induced attractor selection. PLoS ONE 1, e49 (2006).
Loeb, L. A. A mutator phenotype in cancer. Cancer Res. 61, 3230–3239 (2001).
Loeb, L. A., Loeb, K. R. & Anderson, J. P. Multiple mutations and cancer. Proc. Natl Acad. Sci. USA 100, 776–781 (2003).
Tomlinson, I. & Bodmer, W. Selection, the mutation rate and cancer: ensuring that the tail does not wag the dog. Nature Med. 5, 11–12 (1999).
Jablonka, E. Inheritance systems and the evolution of new levels of individuality. J. Theor. Biol. 170, 301–309 (1994).
Keller, A. D. Fixation of epigenetic states in a population. J. Theor. Biol. 176, 211–219 (1995).
Morley, A. Quantifying leukemia. N. Engl. J. Med. 339, 627–629 (1998).
Talpaz, M. et al. Imatinib induces durable hematologic and cytogenetic responses in patients with accelerated phase chronic myeloid leukemia: results of a phase 2 study. Blood 99, 1928–1937 (2002).
Gorre, M. E. et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science 293, 876–880 (2001).
Gambacorti-Passerini, C. B. et al. Molecular mechanisms of resistance to imatinib in Philadelphia-chromosome-positive leukaemias. Lancet Oncol. 4, 75–85 (2003).
Wei, Y. et al. Not all imatinib resistance in CML are BCR-ABL kinase domain mutations. Ann. Hematol. 85, 841–847 (2006).
Okabe, S., Tauchi, T. & Ohyashiki, K. Characteristics of dasatinib- and imatinib-resistant chronic myelogenous leukemia cells. Clin. Cancer Res. 14, 6181–6186 (2008).
Levsky, J. M., Shenoy, S. M., Pezo, R. C. & Singer, R. H. Single-cell gene expression profiling. Science 297, 836–840 (2002).
Cohen, A. A. et al. Dynamic proteomics of individual cancer cells in response to a drug. Science 322, 1511–1516 (2008).
Ptashne, M. On the use of the word 'epigenetic'. Curr. Biol. 17, R233–R236 (2007).
Waddington, C. H. The epigenotype. Endeavour 1, 18–20 (1942).
Aurell, E. & Sneppen, K. Epigenetics as a first exit problem. Phys. Rev. Lett. 88, 048101 (2002).
Walczak, A. M., Onuchic, J. N. & Wolynes, P. G. Absolute rate theories of epigenetic stability. Proc. Natl Acad. Sci. USA 102, 18926–18931 (2005).
Khorasanizadeh, S. The nucleosome: from genomic organization to genomic regulation. Cell 116, 259–272 (2004).
Schaefer, C. B., Ooi, S. K., Bestor, T. H. & Bourc'his, D. Epigenetic decisions in mammalian germ cells. Science 316, 398–399 (2007).
Arney, K. L. & Fisher, A. G. Epigenetic aspects of differentiation. J. Cell Sci. 117, 4355–4363 (2004).
Fuks, F. DNA methylation and histone modifications: teaming up to silence genes. Curr. Opin. Genet. Dev. 15, 490–495 (2005).
Surani, M. A., Hayashi, K. & Hajkova, P. Genetic and epigenetic regulators of pluripotency. Cell 128, 747–762 (2007).
Huang, S. Genomics, complexity and drug discovery: insights from Boolean network models of cellular regulation. Pharmacogenomics 2, 203–222 (2001).
Madhani, H. D. et al. Epigenomics: a roadmap, but to where? Science 322, 43–44 (2008).
Delbrück, M. in Unités biologiques douées de continuité génétique Colloques Internationaux du Centre National de la Recherche Scientifique (CNRS, Paris, 1949).
Monod, J. & Jacob, F. Teleonomic mechanisms in cellular metabolism, growth, and differentiation. Cold Spring Harb. Symp. Quant. Biol. 26, 389–401 (1961).
Kim, K. Y. & Wang, J. Potential energy landscape and robustness of a gene regulatory network: toggle switch. PLoS Comput. Biol. 3, e60 (2007).
Kurchan, J. & Laloux, L. Phase space geometry and slow dynamics. J. Phys. A Math. Gen. 29, 1929–1948 (1996).
Kauffman, S. A. The Origins of Order (Oxford Univ. Press, New York, 1993).
Acknowledgements
S.H. acknowledges support from the Air Force Office of Scientific Research, the National Institutes of Health and the Canada Foundation of Innovation. The authors also thank Alfonso Martinez-Arias for many discussions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Rights and permissions
About this article
Cite this article
Brock, A., Chang, H. & Huang, S. Non-genetic heterogeneity — a mutation-independent driving force for the somatic evolution of tumours. Nat Rev Genet 10, 336–342 (2009). https://doi.org/10.1038/nrg2556
Issue Date:
DOI: https://doi.org/10.1038/nrg2556