Key Points
-
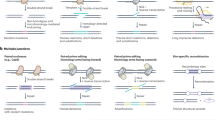
Gene conversion involves the unidirectional transfer of genetic material from a 'donor' to an 'acceptor' sequence. It mediates the transfer of genetic information from intact homologous sequences to regions containing double-strand breaks (DSBs), and can occur between sister chromatids, homologous chromosomes, and even between homologous sequences on the same chromatid or on different chromosomes.
-
Gene conversion occurs predominantly in meiosis, but also in mitosis.
-
Gene conversion seems to result from either synthesis-dependent strand-annealing (SDSA) or double Holliday junction (HJ) dissolution, rather than from the random resolution of double HJs by an HJ resolvase, as predicted by the seminal double-strand break repair (DSBR) model.
-
Although the helicases Srs2, BLM and Rad54 have been reported to promote SDSA individually, the coordinated action of BLM, topoisomerase IIIα and BLAP75 (also known as RMI1) is needed to promote double-HJ dissolution.
-
The rate of gene conversion depends on multiple factors such as sequence homology and distance between the interacting sequences, but might also be affected by the presence of certain specific sequence motifs.
-
In mammalian cells, gene-conversion tracts are usually short and rarely exceed the order of 1 kb in length.
-
Gene-conversion events constitute an important driving force in genome evolution. Although interlocus gene conversion has been implicated in the concerted evolution of many human gene families, interallelic gene conversion has also been found to occur frequently at certain loci, generating a high level of allelic diversity.
-
The impact of gene conversion on the evolution of multigene families has been mediated by several factors, most notably selection.
-
There is growing evidence from both human population genetic studies and sperm typing to show that gene conversion has had an important role in shaping fine-scale patterns of linkage disequilibrium (LD) in the human genome.
-
Given the widespread presence of gene conversion and the short tract length that is usually involved in gene conversion, gene-conversion-generated SNPs often constitute 'holes' in the haplotype blocks that serve to reduce the efficacy of LD-based association studies.
-
High gene-conversion activity is a common feature of both allelic and non-allelic recombination hotspots; at least 25,000 recombination hotspots have been identified across the human genome.
-
An improved understanding of the role of gene conversion in generating or eliminating recombination hotspots in the human genome promises to improve our ability to predict the locations of unstable genomic regions.
-
Gene conversion has been implicated as the cause of various human genetic diseases.
-
Nearly 50% of the donor genes involved in interlocus gene-conversion events that cause human inherited disease are functional or partially functional.
-
Nearly all the disease-causing gene-conversion events resulted in the functional loss of the respective acceptor gene, the sole exception being a 'gain-of-function' mutation that occurred in the serine protease gene PRSS1.
-
Nearly all known cases of disease-causing interlocus gene conversion occurred between highly homologous sequences located on the same chromosome. There is only one example of the acceptor and donor genes involved residing on different chromosomes: the von Willebrand factor gene (VWF) gene (12p13.3) and its pseudogene (22q11.22–q11.23).
-
Our meta-analysis of pathogenic gene-conversion events revealed that, with one exception (88% between the folate receptor 1 gene (FOLR1) and its pseudogene), the homology between the linked donor and the acceptor gene sequences is always >92% and usually >95%.
-
In stark contrast to the frequent detection of pathogenic interlocus gene-conversion events (44 events in 17 genes), the occurrence of interallelic gene-conversion events causing human inherited disease is quite rare, with probably only one bona fide case identified: the SRY (sex determining region Y)-box 9 (SOX9).
-
Only a few well-documented examples of gene-conversion events in cancer have been reported in the literature.
-
The occurrence of de novo gene-conversion events is indicative of the dynamic nature of this genetic process.
-
Gene conversion can account for the occurrence of some recurrent mutations on different chromosomal backgrounds in different ethnic groups.
-
In the future, gene conversion might provide a possible means to bring about 'natural gene therapy' by offering an important alternative to the introduction of an entire functional gene.
Abstract
Gene conversion, one of the two mechanisms of homologous recombination, involves the unidirectional transfer of genetic material from a 'donor' sequence to a highly homologous 'acceptor'. Considerable progress has been made in understanding the molecular mechanisms that underlie gene conversion, its formative role in human genome evolution and its implications for human inherited disease. Here we assess current thinking about how gene conversion occurs, explore the key part it has played in fashioning extant human genes, and carry out a meta-analysis of gene-conversion events that are known to have caused human genetic disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Slightom, J. L., Blechi, A. E. & Smithies, O. Human fetal Gγ- and Aγ-globin genes: complete nucleotide sequences suggest that DNA can be exchanged between these duplicated genes. Cell 21, 627–638 (1980).
Paques, F. & Haber, J. E. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63, 349–404 (1999).
Krogh, B. O. & Symington, L. S. Recombination proteins in yeast. Annu. Rev. Genet. 38, 233–271 (2004).
Szostak, J. W., Orr-Weaver, T. L., Rothstein, R. J. & Stahl, F. W. The double-strand-break repair model for recombination. Cell 33, 25–35 (1983).
Haber, J. E., Ira, G., Malkova, A. & Sugawara, N. Repairing a double-strand chromosome break by homologous recombination: revisiting Robin Holliday's model. Philos. Trans. R. Soc. Lond. B Biol. Sci. 359, 79–86 (2004). This review describes how the seminal DSBR and SDSA models have evolved over time.
Hunter, N. & Kleckner, N. The single-end invasion: an asymmetric intermediate at the double-strand break to double-Holliday junction transition of meiotic recombination. Cell 106, 59–70 (2001).
Ira, G., Satory, D. & Haber, J. E. Conservative inheritance of newly synthesized DNA in double-strand break-induced gene conversion. Mol. Cell. Biol. 26, 9424–9429 (2006).
Ira, G., Malkova, A., Liberi, G., Foiani, M. & Haber, J. E. Srs2 and Sgs1–Top3 suppress crossovers during double-strand break repair in yeast. Cell 115, 401–411 (2003). This study suggests that, whereas Srs2 promotes the SDSA pathway, Sgs1 and the topoisomerase Top3 remove double HJs, both leading to the generation of only gene-conversion events.
Robert, T., Dervins, D., Fabre, F. & Gangloff, S. Mrc1 and Srs2 are major actors in the regulation of spontaneous crossover. EMBO J. 25, 2837–2846 (2006).
Aylon, Y., Liefshitz, B., Bitan-Banin, G. & Kupiec, M. Molecular dissection of mitotic recombination in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 23, 1403–1417 (2003).
Krejci, L. et al. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423, 305–309 (2003).
Veaute, X. et al. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423, 309–312 (2003).
Krejci, L. et al. Role of ATP hydrolysis in the antirecombinase function of Saccharomyces cerevisiae Srs2 protein. J. Biol. Chem. 279, 23193–23199 (2004).
Adams, M. D., McVey, M. & Sekelsky, J. J. Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science 299, 265–267 (2003).
McVey, M., Larocque, J. R., Adams, M. D. & Sekelsky, J. J. Formation of deletions during double-strand break repair in Drosophila DmBLM mutants occurs after strand invasion. Proc. Natl Acad. Sci. USA 101, 15694–15699 (2004).
Weinert, B. T. & Rio, D. C. DNA strand displacement, strand annealing and strand swapping by the Drosophila Bloom's syndrome helicase. Nucleic Acids Res. 35, 1367–1376 (2007).
Bachrati, C. Z., Borts, R. H. & Hickson, I. D. Mobile D-loops are a preferred substrate for the Bloom's syndrome helicase. Nucleic Acids Res. 34, 2269–2279 (2006).
Bugreev, D. V., Mazina, O. M. & Mazin, A. V. Rad54 protein promotes branch migration of Holliday junctions. Nature 442, 590–593 (2006). This work identifies a novel function (that is, promotion of bidirectional DNA branch migration) of the Rad54 protein, and suggests that it could facilitate either the SDSA pathway or the formation of double HJs.
Wu, L. & Hickson, I. D. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature 426, 870–874 (2003). This work shows that BLM and human topoisomerase IIIα suppress crossing over through the mechanism of double-HJ dissolution.
Jessop, L., Rockmill, B., Roeder, G. S. & Lichten, M. Meiotic chromosome synapsis-promoting proteins antagonize the anti-crossover activity of sgs1. PLoS Genet. 2, e155 (2006).
Plank, J. L., Wu. J. & Hsieh, T. S. Topoisomerase IIIα and Bloom's helicase can resolve a mobile double Holliday junction substrate through convergent branch migration. Proc. Natl Acad. Sci. USA 103, 11118–11123 (2006).
Johnson-Schlitz, D. & Engels, W. R. Template disruptions and failure of double Holliday junction dissolution during double-strand break repair in Drosophila BLM mutants. Proc. Natl Acad. Sci. USA 103, 16840–16845 (2006).
Wu, L. et al. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc. Natl Acad. Sci. USA 103, 4068–4073 (2006). This work identifies BLAP75 as the third component of the double-HJ dissolvasome; this finding was confirmed concurrently in reference 24.
Raynard, S., Bussen, W. & Sung, P. A double Holliday junction dissolvasome comprising BLM, topoisomerase IIIα, and BLAP75. J. Biol. Chem. 281, 13861–13864 (2006).
Allers, T. & Lichten, M. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106, 47–57 (2001).
Borner, G. V., Kleckner, N. & Hunter, N. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117, 29–45 (2004).
Liu, Y. & West, S. C. Happy Hollidays: 40th anniversary of the Holliday junction. Nature Rev. Mol. Cell Biol. 5, 937–944 (2004). An historical review of the HJ that remains the basis of our thinking about homologous recombination.
Schildkraut, E., Miller, C. A. & Nickoloff, J. A. Gene conversion and deletion frequencies during double-strand break repair in human cells are controlled by the distance between direct repeats. Nucleic Acids Res. 33, 1574–1580 (2005).
Ezawa, K., Oota, S. & Saitou, N. Genome-wide search of gene conversions in duplicated genes of mouse and rat. Mol. Biol. Evol. 23, 927–940 (2006).
Liskay, R. M., Letsou, A. & Stachelek, J. Homology requirement for efficient gene conversion between duplicated chromosomal sequences in mammalian cells. Genetics 115, 161–167 (1987).
Waldman, A. S. & Liskay, R. M. Dependence of intrachromosomal recombination in mammalian cells on uninterrupted homology. Mol. Cell. Biol. 8, 5350–5357 (1988).
Reiter, L. T. et al. Human meiotic recombination products revealed by sequencing a hotspot for homologous strand exchange in multiple HNPP deletion patients. Am. J. Hum. Genet. 62, 1023–1033 (1998).
Judd, S. R. & Petes, T. D. Physical lengths of meiotic and mitotic gene conversion tracts in Saccharomyces cerevisiae. Genetics 118, 401–410 (1988).
Papadakis, M. N. & Patrinos, G. P. Contribution of gene conversion in the evolution of the human β-like globin gene family. Hum. Genet. 104, 117–125 (1999).
Bosch, E., Hurles, M. E., Navarro, A. & Jobling, M. A. Dynamics of a human interparalog gene conversion hotspot. Genome Res. 14, 835–844 (2004).
Zangenberg, G., Huang, M. M., Arnheim, N. & Erlich, H. New HLA-DPB1 alleles generated by interallelic gene conversion detected by analysis of sperm. Nature Genet. 10, 407–414 (1995).
Jeffreys, A. J. & May, C. A. Intense and highly localized gene conversion activity in human meiotic crossover hot spots. Nature Genet. 36, 151–156 (2004). This work provides evidence for hotspots of human interallelic gene conversion with the potential to exert profound effects on haplotype diversity.
Bacolla, A. et al. Breakpoints of gross deletions coincide with non-B DNA conformations. Proc. Natl Acad. Sci. USA 101, 14162–14167 (2004).
Schildkraut, E., Miller, C. A. & Nickoloff, J. A. Transcription of a donor enhances its use during double-strand break-induced gene conversion in human cells. Mol. Cell. Biol. 26, 3098–3105 (2006).
Gonzalez-Barrera, S., Garcia-Rubio, M. & Aguilera, A. Transcription and double-strand breaks induce similar mitotic recombination events in Saccharomyces cerevisiae. Genetics 162, 603–614 (2002).
Innan, H. A two-locus gene conversion model with selection and its application to the human RHCE and RHD genes. Proc. Natl Acad. Sci. USA 100, 8793–8798 (2003). The results presented here provide an indication of the strength of selection that is required to balance gene conversion in maintaining the observed pattern of allelic variation in this two-locus system.
Hallast, P., Nagirnaja, L., Margus, T. & Laan, M. Segmental duplications and gene conversion: human luteinizing hormone/chorionic gonadotropin β gene cluster. Genome Res. 15, 1535–1546 (2005).
Verrelli, B. C. & Tishkoff, S. A. Signatures of selection and gene conversion associated with human color vision variation. Am. J. Hum. Genet. 75, 363–375 (2004). By means of population genetic and statistical analyses, these authors showed how a combination of natural selection and gene conversion has shaped sequence diversity in the OPN1LW gene.
Sharon, D. et al. Primate evolution of an olfactory receptor cluster: diversification by gene conversion and recent emergence of pseudogenes. Genomics 61, 24–36 (1999).
Woelk, C. H., Frost, S. D., Richman, D. D., Higley, P. E. & Kosakovsky Pond, S. L. Evolution of the interferon α gene family in eutherian mammals. Gene 397, 38–50 (2007).
Plotnikova, O. V. et al. Conversion and compensatory evolution of the γ-crystallin genes and identification of a cataractogenic mutation that reverses the sequence of the human CRYGD gene to an ancestral state. Am. J. Hum. Genet. 81, 32–43 (2007).
Vazquez-Salat, N., Yuhki, N., Beck, T., O'Brien, S. J. & Murphy, W. J. Gene conversion between mammalian CCR2 and CCR5 chemokine receptor genes: a potential mechanism for receptor dimerization. Genomics 90, 213–224 (2007).
Eickbush, T. H. & Eickbush, D. G. Finely orchestrated movements: evolution of the ribosomal RNA genes. Genetics 175, 477–485 (2007).
Hurles, M. E. in: Encyclopedia of Life Sciences (John Wiley & Sons, Chichester, 2003).
Rozen, S. et al. Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature 423, 873–876 (2003). Having estimated that an average of ∼600 nucleotides in each newborn male have undergone Y–Y gene conversion, the authors highlighted an important role for gene conversion in the evolution of multi-copy testis-expressed gene families in the male-specific region of the human Y chromosome.
Hurles, M. E., Willey, D., Matthews, L. & Hussain, S. S. Origins of chromosomal rearrangement hotspots in the human genome: evidence from the AZFa deletion hotspots. Genome Biol. 5, R55 (2004). The authors carried out multiple simulations to explore how gene conversion homogenizes paralogous sequences at the same time that it diversifies orthologous sequences.
Winderickx, J., Battisti, L., Hibiya, Y., Motulsky, A. G. & Deeb, S. S. Haplotype diversity in the human red and green opsin genes: evidence for frequent sequence exchange in exon 3. Hum. Mol. Genet. 2, 1413–1421 (1993).
Carroll, J., Neitz, J. & Neitz, M. Estimates of L:M cone ratio from ERG flicker photometry and genetics. J. Vis. 2, 531–542 (2002).
Joly, E. & Rouillon, V. The orthology of HLA-E and H2-Qa1 is hidden by their concerted evolution with other MHC class I molecules. Biol. Direct 1, 2 (2006).
Yip, S. P. Sequence variation at the human ABO locus. Ann. Hum. Genet. 66, 1–27 (2002).
von Salome, J., Gyllensten, U. & Bergstrom, T. F. Full-length sequence analysis of the HLA-DRB1 locus suggests a recent origin of alleles. Immunogenetics 59, 261–271 (2007).
Hurles, M. Are 100,000 'SNPs' useless? Science 298, 1509 (2002).
Bailey, J. A. et al. Recent segmental duplications in the human genome. Science 297, 1003–1007 (2002).
Fredman, D. et al. Complex SNP-related sequence variation in segmental genome duplications. Nature Genet. 36, 861–866 (2004). One of the first papers to report structural variation in the human genome, providing evidence for a role for gene conversion in promoting the variability of duplicon sequences.
Pavlicek, A., House, R., Gentles, A. J., Jurka, J. & Morrow, B. E. Traffic of genetic information between segmental duplications flanking the typical 22q11.2deletion in velo-cardio-facial syndrome/DiGeorge syndrome. Genome Res. 15, 1487–1495 (2005).
Cheng, Z. et al. A genome-wide comparison of recent chimpanzee and human segmental duplications. Nature 437, 88–93 (2005).
International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature 431, 931–945 (2004).
Jackson, M. S. et al. Evidence for widespread reticulate evolution within human duplicons. Am. J. Hum. Genet. 77, 824–840 (2005).
Ardlie, K. et al. Lower-than-expected linkage disequilibrium between tightly linked markers in humans suggests a role for gene conversion. Am. J. Hum. Genet. 69, 582–589 (2001).
Ptak, S. E., Voelpel, K. & Przeworski, M. Insights into recombination from patterns of linkage disequilibrium in humans. Genetics 167, 387–397 (2004).
International HapMap Consortium. A haplotype map of the human genome. Nature 437, 1299–1320 (2005).
Conrad, D. F. et al. A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nature Genet. 38, 1251–1260 (2006).
de Bakker, P. I. et al. Transferability of tag SNPs in genetic association studies in multiple populations. Nature Genet. 38, 1298–1303 (2006).
Wall, J. D. Close look at gene conversion hot spots. Nature Genet. 36, 114–115 (2004).
Need, A. C. & Goldstein, D. B. Genome-wide tagging for everyone. Nature Genet. 38, 1227–1228 (2006).
Lindsay, S. J., Khajavi, M., Lupski, J. R. & Hurles, M. E. A chromosomal rearrangement hotspot can be identified from population genetic variation and is coincident with a hotspot for allelic recombination. Am. J. Hum. Genet. 79, 890–902 (2006). Together with reference 72, this work reveals the imprint of gene conversion by resequencing mutation-prone disease-associated low copy repeat (LCR) regions (commented on in reference 75).
Raedt, T. D. et al. Conservation of hotspots for recombination in low-copy repeats associated with the NF1 microdeletion. Nature Genet. 38, 1419–1423 (2006).
Myers, S., Bottolo, L., Freeman, C., McVean, G. & Donnelly, P. A fine-scale map of recombination rates and hotspots across the human genome. Science 310, 321–324 (2005).
Coop, G. & Myers, S. R. Live hot, die young: transmission distortion in recombination hotspots. PLoS Genet. 3, e35 (2007).
Myers, S. R. & McCarroll, S. A. New insights into the biological basis of genomic disorders. Nature Genet. 38, 1363–1364 (2006).
Blanco, P. et al. Divergent outcomes of intrachromosomal recombination on the human Y chromosome: male infertility and recurrent polymorphism. J. Med. Genet. 37, 752–758 (2000).
Forbes, S. H., Dorschner, M. O., Le, R. & Stephens, K. Genomic context of paralogous recombination hotspots mediating recurrent NF1 region microdeletion. Genes Chrom. Cancer 41, 12–25 (2004).
Hellenthal, G. & Stephens, M. Insights into recombination from population genetic variation. Curr. Opin. Genet. Dev. 16, 565–572 (2006).
Padhukasahasram, B., Marjoram, P. & Nordborg, M. Estimating the rate of gene conversion on human chromosome 21. Am. J. Hum. Genet. 75, 386–397 (2004).
Frisse, L. et al. Gene conversion and different population histories may explain the contrast between polymorphism and linkage disequilibrium levels. Am. J. Hum. Genet. 69, 831–843 (2001).
Jeffreys, A. J. & Neumann, R. Factors influencing recombination frequency and distribution in a human meiotic crossover hotspot. Hum. Mol. Genet. 14, 2277–2287 (2005).
Holloway, K., Lawson, V. E. & Jeffreys, A. J. Allelic recombination and de novo deletions in sperm in the human β-globin gene region. Hum. Mol. Genet. 15, 1099–1111 (2006).
Marais, G. Biased gene conversion: implications for genome and sex evolution. Trends Genet. 19, 330–338 (2003).
Galtier, N. Gene conversion drives GC content evolution in mammalian histones. Trends Genet. 19, 65–68 (2003).
Spencer, C. C. et al. The influence of recombination on human genetic diversity. PLoS Genet. 2, e148 (2006).
Spencer, C. C. Human polymorphism around recombination hotspots. Biochem. Soc. Trans. 34, 535–536 (2006).
Hernandez, R. D., Williamson, S. H., Zhu, L. & Bustamante, C. D. Context-dependent mutation rates may cause spurious signatures of a fixation bias favoring higher GC-content in humans. Mol. Biol. Evol. 26 Jul 2007 (doi:10.1093/molbev/msm149).
Teich, N. et al. Gene conversion between functional trypsinogen genes PRSS1 and PRSS2 associated with chronic pancreatitis in a six-year-old girl. Hum. Mutat. 25, 343–347 (2005). This paper reports a unique gene-conversion event that occurred between two functional genes, resulting in a gain of function.
Lorson, C. L., Hahnen, E., Androphy, E. J. & Wirth, B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl Acad. Sci. USA 96, 6307–6311 (1999).
Gupta, P. K. et al. Gene conversions are a common cause of von Willebrand disease. Br. J. Haematol. 130, 752–758 (2005).
Linardopoulou, E. V. et al. Human subtelomeres are hot spots of interchromosomal recombination and segmental duplication. Nature 437, 94–100 (2005).
Rudd, M. K. et al. Elevated rates of sister chromatid exchange at chromosome ends. PLoS Genet. 3, e32 (2007).
Lee-Chen, G. J. & Wang, T. R. Mucopolysaccharidosis type I: identification of novel mutations that cause Hurler/Scheie syndrome in Chinese families. J. Med. Genet. 34, 939–941 (1997).
Allen, L. A. et al. A novel loss of function mutation in exon 10 of the FSH receptor gene causing hypergonadotrophic hypogonadism: clinical and molecular characteristics. Hum. Reprod. 18, 251–256 (2003).
Pop, R., Zaragoza, M. V., Gaudette, M., Dahrmann, U. & Scherer, G. A homozygous nonsense mutation in SOX9 in the dominant disorder campomelic dysplasia: a case of mitotic gene conversion. Hum. Genet. 117, 43–53 (2005). This work reports the first fully characterized somatic interallelic gene-conversion event in the context of human inherited disease.
Hauptschein, R. S. et al. An apparent interlocus gene conversion-like event at a putative tumor suppressor gene locus on human chromosome 6q27 in a Burkitt's lymphoma cell line. DNA Res. 7, 261–272 (2000).
Zhang, J. et al. Gene conversion is a frequent mechanism of inactivation of the wild-type allele in cancers from MLH1/MSH2 deletion carriers. Cancer Res. 66, 659–664 (2006).
Auclair, J. et al. Novel biallelic mutations in MSH6 and PMS2 genes: gene conversion as a likely cause of PMS2 gene inactivation. Hum. Mutat. 7 Jun 2007 (doi:10.humu.20569).
Chambers, S. R., Hunter, N., Louis, E. J. & Borts, R. H. The mismatch repair system reduces meiotic homeologous recombination and stimulates recombination-dependent chromosome loss. Mol. Cell. Biol. 16, 6110–6120 (1996).
Romano, M. et al. Regulation of 3′ splice site selection in the 844ins68 polymorphism of the cystathionine β-synthase gene. J. Biol. Chem. 277, 43821–43829 (2002).
Vyletal, P. et al. Haplotype diversity of cystathionine β-synthase alleles bearing the most common homocystinuria mutation c.833T>C: a possible role for gene conversion. Hum. Mutat. 28, 255–264 (2007).
Jonkman, M. F. et al. Revertant mosaicism in epidermolysis bullosa caused by mitotic gene conversion. Cell 88, 543–551 (1997).
Ogino, S., Gao, S., Leonard, D. G., Paessler, M. & Wilson, R. B. Inverse correlation between SMN1 and SMN2 copy numbers: evidence for gene conversion from SMN2 to SMN1. Eur. J. Hum. Genet. 11, 275–277 (2003).
Fichou, Y. & Férec, C. The potential of oligonucleotides for therapeutic applications. Trends Biotechnol. 24, 563–570 (2006).
Shinohara, A., Ogawa, H. & Ogawa, T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell 69, 457–470 (1992).
Masson, J. Y. et al. Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes Dev. 15, 3296–3307 (2001).
Liu, Y., Masson, L. Y., Shah, R., O'Regan, P. & West, S. C. RAD51C is required for Holliday junction processing in mammalian cells. Science 303, 243–246 (2004).
Lisby, M. & Rothstein, R. DNA repair: keeping it together. Curr. Biol. 14, R994–R996 (2004).
De Jager, M. et al. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol. Cell 8, 1129–1135 (2001).
Moreno-Herrero, F. et al. Mesoscale conformational changes in the DNA-repair complex Rad50/Mre11/Nbs1 upon binding DNA. Nature 437, 440–443 (2005).
Scully, R. et al. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell 88, 265–275 (1997).
Sharan, S. K. et al. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking BRCA2. Nature 386, 804–810 (1997).
Chen, J. et al. Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol. Cell 2, 317–328 (1998).
Yuan, S. S. et al. BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res. 59, 3547–3551 (1999).
Esashi, F., Galkin, V. E., Yu, X., Egelman, E. H. & West, S. C. Stabilization of RAD51 nucleoprotein filaments by the C-terminal region of BRCA2. Nature Struct. Mol. Biol. 14, 468–474 (2007).
Davies, O. R. & Pellegrini, L. Interaction with the BRCA2 C terminus protects RAD51–DNA filaments from disassembly by BRC repeats. Nature Struct. Mol. Biol. 14, 475–483 (2007).
Roy, A. M. et al. Potential gene conversion and source genes for recently integrated Alu elements. Genome Res. 10, 1485–1495 (2000).
Zhi, D. Sequence correlation between neighboring Alu instances suggests post-retrotransposition sequence exchange due to Alu gene conversion. Gene 390, 117–121 (2007).
Sen, S. K. et al. Human genomic deletions mediated by recombination between Alu elements. Am. J. Hum. Genet. 79, 41–53 (2006).
Tremblay, A., Jasin, M. & Chartrand, P. A double-strand break in a chromosomal LINE element can be repaired by gene conversion with various endogenous LINE elements in mouse cells. Mol. Cell. Biol. 20, 54–60 (2000).
Wurtele, H., Gusew. N., Lussier, R. & Chartrand, P. Characterization of in vivo recombination activities in the mouse embryo. Mol. Genet. Genomics 273, 252–263 (2005).
Myers, J. S. et al. A comprehensive analysis of recently integrated human Ta L1 elements. Am. J. Hum. Genet. 71, 312–326 (2002).
Vincent, B. J. et al. Following the LINEs: an analysis of primate genomic variation at human-specific LINE-1 insertion sites. Mol. Biol. Evol. 20, 1338–1348 (2003).
Chen, J. M., Férec, C. & Cooper, D. N. Mechanism of Alu integration into the human genome. Genome Med. 1, 9–17 (2007).
Khan, H., Smit, A. & Boissinot, S. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res. 16, 78–87 (2006).
Maizels, N. Immunoglobulin gene diversification. Annu. Rev. Genet. 39, 23–46 (2005).
Tang, E. S. & Martin, A. Immunoglobulin gene conversion: Synthesizing antibody diversification and DNA repair. DNA Repair 27 Jun 2007 (doi:10.1016/j.dnarep.2007.05.002).
D'Avirro, N., Truong, D., Xu, B. & Selsing, E. Sequence transfers between variable regions in a mouse antibody transgene can occur by gene conversion. J. Immunol. 175, 8133–8137 (2005).
Darlow, J. M. & Stott, D. I. Gene conversion in human rearranged immunoglobulin genes. Immunogenetics 58, 511–522 (2006).
Hayakawa, T. et al. A human-specific gene in microglia. Science 309, 1693 (2005). This work describes a human-specific gene-conversion event that might have been significant in the evolution of the Homo genus.
Heinen, S. et al. De novo gene conversion in the RCA gene cluster (1q32) causes mutations in complement factor H associated with atypical hemolytic uremic syndrome. Hum. Mutat. 27, 292–293 (2006).
Lee, H. H., Tsai, F. J., Lee, Y. J. & Yang, Y. C. Diversity of the CYP21A2 gene: a 6.2-kb TaqI fragment and a 3.2-kb TaqI fragment mistaken as CYP21A1P. Mol. Genet. Metab. 88, 372–277 (2006).
Friaes, A. et al. CYP21A2 mutations in Portuguese patients with congenital adrenal hyperplasia: identification of two novel mutations and characterization of four different partial gene conversions. Mol. Genet. Metab. 88, 58–65 (2006).
Higashi, Y., Tanae, A., Inoue, H., Hiromasa, T. & Fujii-Kuriyama, Y. Aberrant splicing and missense mutations cause steroid 21-hydroxylase [P-450(C21)] deficiency in humans: possible gene conversion products. Proc. Natl Acad. Sci. USA 85, 7486–7490 (1988).
Fardella, C. E. et al. Gene conversion in the CYP11B2 gene encoding P450c11AS is associated with, but does not cause, the syndrome of corticosterone methyloxidase II deficiency. J. Clin. Endocrinol. Metab. 81, 321–326 (1996).
Nicod, J., Dick, B., Frey, F. J. & Ferrari, P. Mutation analysis of CYP11B1 and CYP11B2 in patients with increased 18-hydroxycortisol production. Mol. Cell. Endocrinol. 214, 167–174 (2004).
Sarhadi, V. et al. A unique form of autosomal dominant cataract explained by gene conversion between β-crystallin B2 and its pseudogene. J. Med. Genet. 38, 392–396 (2001).
De Marco, P. et al. Folate pathway gene alterations in patients with neural tube defects. Am. J. Med. Genet. 95, 216–223 (2000).
Hatton, C. E., Cooper, A., Whitehouse, C. & Wraith, J. E. Mutation analysis in 46 British and Irish patients with Gaucher's disease. Arch. Dis. Child. 77, 17–22 (1997).
Latham, T., Grabowski, G. A., Theophilus, B. D. & Smith, F. I. Complex alleles of the acid β-glucosidase gene in Gaucher disease. Am. J. Hum. Genet. 47, 79–86 (1990).
Eyal, N., Wilder, S. & Horowitz, M. Prevalent and rare mutations among Gaucher patients. Gene 96, 277–283 (1990).
Hong, C. M., Ohashi, T., Yu, X. J., Weiler, S. & Barranger, J. A. Sequence of two alleles responsible for Gaucher disease. DNA Cell Biol. 9, 233–241 (1990).
Millar, D. S. et al. Novel mutations of the growth hormone 1 (GH1) gene disclosed by modulation of the clinical selection criteria for individuals with short stature. Hum. Mutat. 21, 424–440 (2003).
Adams, J. G. III, Marrison, W. T. & Steinberg, M. H. Hemoglobin Parchman: double crossover within a single human gene. Science 218, 291–293 (1982).
Patrinos, G. P. et al. The Cretan type of non-deletional hereditary persistence of fetal hemoglobin [Aγ −158 C>T] results from two independent gene conversion events. Hum. Genet. 102, 629–634 (1998).
Minegishi, Y. et al. Mutations in the human λ5/14.1gene results in B cell deficiency and agammaglobulinemia. J. Exp. Med. 187, 71–77 (1998).
Roesler, J. et al. Recombination events between the p47-phox gene and its highly homologous pseudogenes are the main cause of autosomal recessive chronic granulomatous disease. Blood 95, 2150–2156 (2000).
Vazquez, N. et al. Mutational analysis of patients with p47-phox-deficient chronic granulomatous disease: the significance of recombination events between the p47-phox gene (NCF1) and its highly homologous pseudogenes. Exp. Hematol. 29, 234–243 (2001).
Reyniers, E. et al. Gene conversion between red and defective green opsin gene in blue cone monochromacy. Genomics 29, 323–328 (1995).
Watnick, T. J., Gandolph, M. A., Weber, H., Neumann, H. P. & Germino, G. G. Gene conversion is a likely cause of mutation in PKD1. Hum. Mol. Genet. 7, 1239–1243 (1998).
Inoue, S. et al. Mutation analysis in PKD1 of Japanese autosomal dominant polycystic kidney disease patients. Hum. Mutat. 19, 622–628 (2002).
Nicolis, E., Bonizzato, A., Assael, B. M. & Cipolli, M. Identification of novel mutations in patients with Shwachman–Diamond syndrome. Hum. Mutat. 25, 410 (2005).
Nakashima, E. et al. Novel SBDS mutations caused by gene conversion in Japanese patients with Shwachman–Diamond syndrome. Hum. Genet. 114, 345–348 (2004).
Boocock, G. R. et al. Mutations in SBDS are associated with Shwachman–Diamond syndrome. Nature Genet. 33, 97–101 (2003).
Bussaglia, E. et al. A frame-shift deletion in the survival motor neuron gene in Spanish spinal muscular atrophy patients. Nature Genet. 11, 335–337 (1995).
Eikenboom, J. C., Castaman, G., Vos, H. L., Bertina, R. M. & Rodeghiero, F. Characterization of the genetic defects in recessive type 1 and type 3 von Willebrand disease patients of Italian origin. Thromb. Haemost. 79, 709–717 (1998).
Eikenboom, J. C., Vink, T., Briët, E., Sixma, J. J. & Reitsma, P. H. Multiple substitutions in the von Willebrand factor gene that mimic the pseudogene sequence. Proc. Natl Acad. Sci. USA 91, 2221–2224 (1994).
Zhang, Z. P., Blombäck, M., Nyman, D. & Anvret, M. Mutations of von Willebrand factor gene in families with von Willebrand disease in the Aland Islands. Proc. Natl Acad. Sci. USA 90, 7937–7940 (1993).
Holmberg, L. et al. von Willebrand factor mutation enhancing interaction with platelets in patients with normal multimeric structure. J. Clin. Invest. 91, 2169–2177 (1993).
Surdhar, G. K., Enayat, M. S., Lawson, S., Williams, M. D. & Hill, F. G. Homozygous gene conversion in von Willebrand factor gene as a cause of type 3 von Willebrand disease and predisposition to inhibitor development. Blood 98, 248–250 (2001).
Acknowledgements
We regret that, owing to space limitations, much of the relevant work could not be cited. We are indebted to R. Kanaar for critically reading the manuscript and for his valuable and detailed comments. We also thank the three anonymous reviewers for their constructive criticism of the manuscript. This work was partially supported by the INSERM (Institut National de la Santé et de la Recherche Médicale), France, and the Erasmus University Medical Center, Rotterdam, The Netherlands.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information S1
Collation of human pathogenic gene–conversion events (PDF 176 kb)
Related links
Glossary
- Unequal crossover
-
A recombination event between non-allelic sequences on non-sister chromatids of a pair of homologous chromosomes.
- Homologous recombination
-
The process by which segments of DNA are exchanged between two DNA duplexes that share high sequence similarity.
- Double-strand break
-
Breaks in opposite DNA strands that lie within ∼10–20 bp of each other.
- Holliday junction
-
A point at which the strands of two dsDNA molecules exchange partners, an event that occurs as an intermediate in crossing over or gene conversion.
- Mismatch repair
-
A natural enzymatic process that replaces a mispaired nucleotide within a DNA duplex to yield perfect Watson–Crick base pairing.
- Orthologue
-
A homologous gene that is derived from a speciation event or by vertical descent.
- Gene-conversion tract
-
In theory, this is the portion of the 'acceptor' sequence that is copied from the 'donor'. Because in practice the length of the tract cannot be precisely known, it must be expressed in terms of the lengths of the minimal and maximal converted tracts: the former refers to the entire region spanned by converted discriminant nucleotides and the latter refers to the region that is delimited by the two nearest unconverted discriminant nucleotides between the donor and acceptor sequences.
- Double crossover
-
Two crossovers that occur in a chromosomal region between highly homologous genes, resulting in reciprocal sequence exchange between them.
- Chi (χ) sequence
-
An 8-bp sequence (5′-GCTGGTGG-3′) that acts as a recombination hotspot in Escherichia coli.
- Z-DNA
-
One of several possible double-helical structures of DNA. Z-DNA is a left-handed double-helical structure in which the double helix winds to the left in a zig-zag pattern, rather than to the right, as occurs in the more common B-DNA form.
- Concerted evolution
-
The process by which repetitive DNA sequences are homogenized such that the individual members of a given DNA repeat or multigene family in one species come to show a higher degree of sequence identity with each other than they do with members of the same DNA repeat or multigene family in another species.
- Paralogue
-
One of a set of homologous genes in the same species that have evolved from a gene duplication, and that can be associated with a subsequent divergence of function.
- Segmental duplication
-
A segment of DNA of larger than 1 kb that occurs in two or more copies per haploid genome, with the different copies sharing >90% sequence identity.
- Linkage disequilibrium
-
A statistical association between particular alleles at two or more neighbouring loci on the same chromosome that results from a specific ancestral haplotype being common in the population under study.
- Tag SNPs
-
SNPs that are correlated with, and can therefore serve as a proxy for, much of the known remaining common variation in a region.
- Somatic hypermutation
-
A process that occurs after immunoglobulin gene rearrangement, whereby the base sequences of part of the immunoglobulin variable regions are mutated more frequently than the rest of the genome. This sequence variation is subject to a selection process in the immune system that favours those cells that express immunoglobulins with the highest affinity for an antigen.
- Class switch recombination
-
The somatic recombination process by which immunoglobulin isotypes are switched to IgG, IgA or IgE, without altering antigen specificity.
- Multiplex ligation-dependent probe amplification analysis
-
A semi-quantitative PCR-based method that allows multiple targets to be amplified with only a single primer pair, and that is widely used for detecting copy-number variations.
Rights and permissions
About this article
Cite this article
Chen, JM., Cooper, D., Chuzhanova, N. et al. Gene conversion: mechanisms, evolution and human disease. Nat Rev Genet 8, 762–775 (2007). https://doi.org/10.1038/nrg2193
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg2193
This article is cited by
-
Unexpectedly high mutation rate of cyp11b1 compared to cyp21a2 in randomly-selected turkish women: a large screening study
Journal of Endocrinological Investigation (2023)
-
Limitations of gene editing assessments in human preimplantation embryos
Nature Communications (2023)
-
Conversion between duplicated genes generated by polyploidization contributes to the divergence of poplar and willow
BMC Plant Biology (2022)
-
Supergene origin and maintenance in Atlantic cod
Nature Ecology & Evolution (2022)
-
Genome doubling enabled the expansion of yeast vesicle traffic pathways
Scientific Reports (2022)