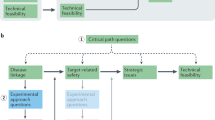
Key Points
-
Academic drug discovery offers an opportunity to effectively harness curiosity-driven research to improve human and animal health, but it is not without risk.
-
We believe that the associated risks can be managed by considering at least five factors that affect the success or failure of projects: organization, target selection, assay design, medicinal chemistry and preclinical pharmacology.
-
This manuscript presents guidelines for reducing the risk that can be caused by poor planning in any of these areas.
Abstract
The number of academic drug discovery centres has grown considerably in recent years, providing new opportunities to couple the curiosity-driven research culture in academia with rigorous preclinical drug discovery practices used in industry. To fully realize the potential of these opportunities, it is important that academic researchers understand the risks inherent in preclinical drug discovery, and that translational research programmes are effectively organized and supported at an institutional level. In this article, we discuss strategies to mitigate risks in several key aspects of preclinical drug discovery at academic drug discovery centres, including organization, target selection, assay design, medicinal chemistry and preclinical pharmacology.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Frye, S., Crosby, M., Edwards, T. & Juliano, R. US academic drug discovery. Nature Rev. Drug Discov. 10, 409–410 (2011). This is a comprehensive overview of the state and challenges of US academic drug discovery pre-2011.
Abou-Gharbia, M. & Childers, W. E. Discovery of innovative therapeutics: today's realities and tomorrow's vision. 2. Pharma's challenges and their commitment to innovation. J. Med. Chem. 57, 5525–5553 (2014).
Khanna, I. Drug discovery in pharmaceutical industry: productivity challenges and trends. Drug Discov. Today 17, 1088–1102 (2012).
Zoghbi, H. Y. The basics of translation. Science 339, 250 (2013).
Gurvich, V. J. & Byrn, S. R. NIPTE: a multi-university partnership supporting academic drug development. Drug Discov. Today 18, 916–921 (2013).
Hasson, S. & Inglese, J. Innovation in academic chemical screening: filling the gaps in chemical biology. Curr. Opin. Chem. Biol. 17, 329–338 (2013).
Dosa, P. I. et al. From HTS to Phase I: the Institute for Therapeutics Discovery and Development at the University of Minnesota. Comb. Chem. High Throughput Screen. 17, 231–240 (2014).
Alberts, B., Kirschner, M., Tilghman, S. & Varmus, H. Rescuing US biomedical research from its systemic flaws. Proc. Natl Acad. Sci. USA 111, 5773–5777 (2014).
Macdonald, G. J. & Lindsley, C. W. A unique industrial–academic collaboration towards the next generation of schizophrenia therapeutics. Curr. Top. Med. Chem. 14, 304–312 (2014).
Munos, B. H. & Chin, W. W. How to revive breakthrough innovation in the pharmaceutical industry. Sci. Transl. Med. 3, 1–3 (2011). This paper argues for high-risk research as an avenue towards breakthrough drugs, and examines the conflict between the rewards of pioneering work and the failure of profit-motivated research and development.
Paul, S. M. et al. How to improve R&D productivity: the pharmaceutical industry's grand challenge. Nature Rev. Drug Discov. 9, 203–214 (2010).
Mackay, M., Street, S. D. A. & McCall, J. M. Risk reduction in drug discovery and development. Curr. Top. Med. Chem. 5, 1087–1090 (2005).
Huryn, D. M. Drug discovery in an academic setting: playing to the strengths. ACS Med. Chem. Lett. 4, 313–315 (2013). This paper details the unique strengths of academic drug discovery: the ability to pursue high-risk projects and the depth of expertise across a broad range of scientific fields.
Ungar, T. & Marcus, M. The innovation forager: stimulating academic innovation. Acad. Med. 89, 194 (2014).
Jorgensen, W. L. Challenges for academic drug discovery. Angew. Chem. Int. Ed. Engl. 51, 11680–11684 (2012).
Ashlock, M. A. & Olson, E. R. Therapeutics development for cystic fibrosis: a successful model for a multisystem genetic disease. Annu. Rev. Med. 62, 107–125 (2011).
Cuatrecasas, P. Drug discovery in jeopardy. J. Clin. Invest. 116, 2837–2842 (2006).
Bruneel, J., D'Este, P. & Salter, A. Investigating the factors that diminish the barriers to university–industry collaboration. Res. Policy 39, 858–868 (2010).
Inglese, J. et al. Genome editing-enabled HTS assays expand drug target pathways for Charcot–Marie-tooth disease. ACS Chem. Biol. 9, 2594–2602 (2014).
Plenge, R. M., Scolnick, E. M. & Altshuler, D. Validating therapeutic targets through human genetics. Nature Rev. Drug Discov. 12, 581–594 (2013). This review evaluates the methods used for drug target validation.
Robertson, G. Towards a more robust approach to selecting and prosecuting promising targets and compounds. Future Med. Chem. 2, 25–34 (2010).
Yan, C. & Higgins, P. J. Drugging the undruggable: transcription therapy for cancer. Biochim. Biophys. Acta 1835, 76–85 (2013).
Rual, J.-F. et al. Towards a proteome-scale map of the human protein–protein interaction network. Nature 437, 1173–1178 (2005).
Rask-Andersen, M., Almén, M. S. & Schiöth, H. B. Trends in the exploitation of novel drug targets. Nature Rev. Drug Discov. 10, 579–590 (2011).
Strachan, R. T., Ferrara, G. & Roth, B. L. Screening the receptorome: an efficient approach for drug discovery and target validation. Drug Discov. Today 11, 706–716 (2006).
Gashaw, I., Ellinghaus, P., Sommer, A. & Asadullah, K. What makes a good drug target? Drug Discov. Today 17, S24–S30 (2012). This article provides a pharmaceutical industry description of the evaluation of drug targets.
Peers, I. S., South, M. C., Ceuppens, P. R., Bright, J. D. & Pilling, E. Can you trust your animal study data? Nature Rev. Drug Discov. 13, 560 (2014).
Smith, M. A. & Houghton, P. A. Proposal regarding reporting of in vitro testing results. Clin. Cancer Res. 19, 2828–2833 (2013).
Prinz, F., Schlange, T. & Asadullah, K. Believe it or not: how much can we rely on published data on potential drug targets? Nature Rev. Drug Discov. 10, 712–713 (2011).
Surade, S. & Blundell, T. L. Structural biology and drug discovery of difficult targets: the limits of ligandability. Chem. Biol. 19, 42–50 (2012).
Egner, U. & Hillig, R. C. A structural biology view of target drugability. Exp. Opin. Drug Discov. 3, 391–401 (2008).
Perola, E., Herman, L. & Weiss, J. Development of a rule-based method for the assessment of protein druggability. J. Chem. Inf. Model. 52, 1027–1038 (2012).
Herschel, M. Portfolio decisions in early development: don't throw out the baby with the bathwater. Pharm. Med. 26, 77–84 (2012).
Vidalin, O., Muslmani, M., Estienne, C., Echchakir, H. & Abina, A. M. In vivo target validation using gene invalidation, RNA interference and protein functional knockout models: it is the time to combine. Curr. Opin. Pharmacol. 9, 669–676 (2009).
Wyatt, P. G., Gilbert, I. H., Read, K. D. & Fairlamb, A. H. Target validation: linking target and chemical properties to desired product profile. Curr. Topics Med. Chem. 11, 1275–1283 (2011).
Bunnage, M. E., Chekler, E. L. P. & Jones, L. H. Target validation using chemical probes. Nature Chem. Biol. 9, 195–199 (2013).
Decher, N., Netter, M. F. & Streit, A. K. Putative impact of RNA editing on drug discovery. Chem. Biol. Drug Des. 81, 13–21 (2013).
Vandamme, D., Minke, B. A., Fitzmaurice, W., Kholodenko, B. N. & Kolch, W. Systems biology-embedded target validation: improving efficacy in drug discovery. Wiley Interdiscip. Rev. Syst. Biol. Med. 6, 1–11 (2014).
Cucurull-Sanchez, L., Spink, K. G. & Moschos, S. A. Relevance of systems pharmacology in drug discovery. Drug Discov. Today 17, 665–670 (2012).
Fishman, M. C. Power of rare diseases: found in translation. Sci. Transl. Med. 201, 201ps11 (2013). This article describes examples from small cohort, first-in-human clinical studies, involving mechanistically homogeneous patient groups that have a genetic basis for a disease, as proof-of-concept trials.
Wendler, A. & Wehling, M. The translatability of animal models for clinical development: biomarkers and disease models. Curr. Opin. Pharmacol. 10, 601–606 (2010).
Wanner, J., Fry, D. C., Peng, Z. & Roberts, J. Druggability assessment of protein–protein interfaces. Future Med. Chem. 3, 2021–2038 (2011).
Kozakov, D. et al. Structural conservation of druggable hot spots in protein–protein interfaces. Proc. Natl Acad. Sci. USA 108, 13528–13533 (2011).
Fuller, J. C., Burgoyne, N. J. & Jackson, R. M. Predicting druggable binding sites at the protein–protein interface. Drug Discov. Today 14, 155–161 (2009).
Lee, J. A. & Berg, E. L. Neoclassic drug discovery: the case for lead generation using phenotypic and functional approaches. J. Biomol. Screen. 18, 1143–1155 (2013).
Sams-Dodd, F. Target-based drug discovery: is something wrong? Drug Discov. Today 10, 139–147 (2005).
Swinney, D. Phenotypic versus target-based drug discovery for first-in-class medicines. Clin. Pharm. Ther. 93, 299–301 (2013). This article promotes phenotypic assays for the identification of first-in-class drugs from which the mechanism of action can be determined.
Welch, E. et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature 447, 87–91 (2007).
Auld, D. S. et al. Molecular basis for the high-affinity binding and stabilization of firefly luciferase by PTC124. Proc. Natl Acad. Sci. USA 107, 4878–4883 (2010).
McElroy, S. P. et al. A lack of premature termination codon read-through efficacy of PTC124 (Ataluren) in a diverse array of reporter assays PLOS Biol. 11, e1001593 (2013).
Loregian, A. & Palù, G. How academic labs can approach the drug discovery process as a way to synergize with big pharma. Trends Microbiol. 21, 261–264 (2013).
DeWoskin, V. A. & Million, R. P. The epigenetics pipeline. Nature Rev. Drug Discov. 12, 661–662 (2013).
Chugh, R. et al. A preclinical evaluation of Minnelide as a therapeutic agent against pancreatic cancer. Sci. Transl. Med. 4, 156ra139 (2012).
Sittampalam, G. S. et al. Assay Guidance Manual http://www.ncbi.nlm.nih.gov/books/NBK53196/ (National Institutes of Health, 2004).
Thorne, N., Auld, D. S. & Inglese, J. Apparent activity in high-throughput screening: origins of compound-dependent assay interference. Curr. Opin. Chem. Biol. 14, 315–324 (2010). This is a comprehensive evaluation of potential interference mechanisms that are commonly encountered in high-throughput screening.
Baell, J. B. & Holloway, G. A. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 53, 2719–2740 (2010). This is the seminal manuscript on pan-assay interference compounds.
Inglese, J. et al. High-throughput screening assays for the identification of chemical probes. Nature Chem. Biol. 3, 466–479 (2007).
Hasson, S. A. et al. Chemogenomic profiling of endogenous PARK2 expression using a genome-edited coincidence reporter. ACS Chem. Biol. http://doi:10.1021/cb5010417 (2015).
Feng, B. Y. & Shoichet, B. K. A detergent-based assay for the detection of promiscuous inhibitors. Nature Protoc. 1, 550–553 (2006). This paper describes an important diagnostic assay from the laboratory that first defined a major source of HTS artifacts associated with the colloidal aggregates many library compounds form in assay buffers.
Lushington, G. & Chaguturu, R. To screen or not to screen: an impassioned plea for smarter chemical libraries to improve drug lead finding. Future Med. Chem. 6, 497–502 (2014).
Baell, J. B. Broad coverage of commercially available lead-like screening space with fewer than 350,000 compounds. J. Chem. Inf. Model. 53, 39–55 (2013).
Matson, S. L. et al. Best practices in compound management for preserving compound integrity and accurately providing samples for assays. J. Biomol. Screen. 14, 476–484 (2009).
Lovering, F., Bikker, J. & Humblet, C. Escape from Flatland: increasing saturation as an approach to improving clinical success. J. Med. Chem. 52, 6752–6756 (2009).
Schreiber, S. L. Organic synthesis toward small-molecule probes and drugs. Proc. Natl Acad. Sci. USA 108, 6699–6702 (2011).
Hergenrother, P. J. Obtaining and screening compound collections: a user's guide and a call to chemists. Curr. Opin. Cell Biol. 10, 213–218 (2006).
Bruns, R. F. & Watson, I. A. Rules for identifying potentially reactive or promiscuous compounds. J. Med. Chem. 55, 9763–9772 (2012).
Walters, W. P. & Namchuk, M. A guide to drug discovery: designing screens: how to make your hits a hit. Nature Rev. Drug Discov. 2, 259–266 (2003).
Baell, J. & Walters, M. A. Chemistry: chemical con artists foil drug discovery. Nature 513, 481–483 (2014).
Saubern, S., Guha, R. & Baell, J. B. KNIME workflow to assess PAINS filters in SMARTS Format. Comparison of RDKit and Indigo cheminformatics libraries. Mol. Inform. 30, 847–850 (2011).
Han, L., Wang, Y. & Bryant, S. H. A survey of across-target bioactivity results of small molecules in PubChem. Bioinformatics 25, 2251–2255 (2009).
Huth, J. R. et al. ALARM NMR: a rapid and robust experimental method to detect reactive false positives in biochemical screens. J. Am. Chem. Soc. 127, 217–224 (2005).
Yang, J. et al. BioActivity Data Associative Promiscuity Pattern Learning Engine http://pasilla.health.unm.edu/tomcat/badapple/badapple (2014).
Dahlin, J. L. & Walters, M. A. The essential roles of chemistry in high-throughput screening triage. Future. Med. Chem. 6, 1265–1290 (2014).
Inglese, J. et al. Quantitative high-throughput screening: a titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc. Natl Acad. Sci. USA 103, 11473–11478 (2006).
Lyssiotis, C. et al. Reprogramming of murine fibroblasts to induced pluripotent stem cells with chemical complementation of Klf4. Proc. Natl Acad. Sci. USA 106, 8912–8917 (2009).
Jacob, N. T., Lockner, J. W., Kravchenko, V. V. & Janda, K. D. Pharmacophore reassignment for induction of the immunosurveillance cytokine TRAIL. Angew. Chem. Int. Ed. 53, 6628–6631 (2014).
Hermann, J. C. et al. Metal impurities cause false positives in high-throughput screening campaigns. ACS Med. Chem. Lett. 4, 197–200 (2013).
Kenakin, T. Quantifying biological activity in chemical terms: a pharmacology primer to describe drug effect. ACS Chem. Biol. 4, 249–260 (2009). This article reviews the fundamental principles of pharmacology that are essential to those involved in any aspect of drug discovery and development.
Baell, J. B. Observations on screening-based research and some concerning trends in the literature. Future Med. Chem. 2, 1529–1546 (2010).
Chung, C. et al. Discovery and characterization of small molecule inhibitors of the BET family bromodomains. J. Med. Chem. 54, 3827–3838 (2011).
Lessene, G. et al. Structure-guided design of a selective BCL-XL inhibitor. Nature Chem. Biol. 9, 390–397 (2013).
Peters, J.-U. et al. Can we discover pharmacological promiscuity early in the drug discovery process? Drug Discov. Today 17, 325–335 (2012).
Molina, D. M. et al. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 341, 84–87 (2013). This paper describes a highly enabling method to determine the extent to which a compound binds to a target protein by virtue of its capacity to stabilize the target protein from thermal denaturation.
Jafari, R. et al. The cellular thermal shift assay for evaluating drug target interactions in cells. Nature Protoc. 9, 2100–2122 (2014).
Lee, J. C. et al. A protein kinase involved in the regulation of inflammatory cytokine biosynthesis. Nature 372, 739–746 (1994). This is a harbinger of the power of phenotypic assays in what was then the new era of molecular biology.
Bollag, G. et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature 467, 596–599 (2010).
Workman, P. & Collins, I. Probing the probes: fitness factors for small molecule tools. Chem. Biol. 17, 561–577 (2010). This is a comprehensive description of the quality attributes that define good probes, leads and drugs.
Kerns, E. H. & Di, L. Drug-like Properties: Concepts, Structure Design and Methods: from ADME to Toxicity Optimization (Academic Press, 2008).
Frearson, J. A. et al. N-myristoyltransferase inhibitors as new leads to treat sleeping sickness. Nature 464, 728–732 (2010). This paper contains an excellent example of a SAR correlation between a recombinant molecular target and parasite proliferation.
Nwaka, S. & Ridley, R. G. Science and society: virtual drug discovery and development for neglected diseases through public–private partnerships. Nature Rev. Drug Discov. 2, 919–928 (2003).
Silber, B. M. Driving drug discovery: the fundamental role of academic labs. Sci. Transl. Med. 2, 30cm16 (2010).
Abulwerdi, F. et al. A novel small-molecule inhibitor of Mcl-1 blocks pancreatic cancer growth in vitro and in vivo. Mol. Cancer Ther. 13, 565–575 (2014).
Ge, Y. et al. Discovery and synthesis of hydronaphthoquinones as novel proteasome inhibitors. J. Med. Chem. 55, 1978–1998 (2012).
Qin, J. et al. Identification of a novel family of BRAFV600E inhibitors. J. Med. Chem. 55, 5220–5230 (2012).
Zhuang, C., Narayanapillai, S., Zhang, W., Sham, Y. Y. & Xing, C. Rapid identification of Keap1-Nrf2 small-molecule inhibitors through structure-based virtual screening and hit-based substructure search. J. Med. Chem. 57, 1121–1126 (2014).
Dahlin, J. et al. PAINS in the assay: chemical mechanisms of assay interference and promiscuous enzymatic inhibition observed during a sulfhydryl-scavenging HTS. J. Med. Chem. http://dx.doi.org/10.1021/jm5019093 (2015).
Sinko, W. et al. Undecaprenyl diphosphate synthase inhibitors: antibacterial drug leads. J. Med. Chem. 57, 5693–5701 (2014).
Johnson, S. et al. A biochemical screen for GroEL/GroES inhibitors. Bioorg. Med. Chem. Lett. 24, 786–789 (2014).
Yan, D. et al. Dual myxovirus screen identifies a small-molecule agonist of the host antiviral response. J. Virol. 87, 11076–11087 (2013).
Ingólfsson, H. I. et al. Phytochemicals perturb membranes and promiscuously alter protein function. ACS Chem. Biol. 9, 1788–1798 (2014).
Kadam, A. et al. Development of novel pyrazolone derivatives as inhibitors of aldose reductase: an eco-friendly one-pot synthesis, experimental screening and in silico analysis. Bioorg. Chem. 53, 67–74 (2014).
Schonbrunn, E. et al. Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases. J. Med. Chem. 56, 3768–3782 (2013).
Acknowledgements
J.L.D. was supported by the following funding bodies: an NIH predoctoral fellowship (F30 DK092026-01); a Pharmaceutical Research and Manufacturers of America Foundation predoctoral pharmacology/toxicology fellowship; and the Mayo Foundation. J.I. is supported by the intramural programme in the National Center for Advancing Translational Sciences (NCATS), at the NIH. M.A.W. acknowledges research funding from the NIH and the Minnesota Partnership for Biotechnology and Medical Genomics. The authors specifically acknowledge the helpful comments and corrections suggested by the referees. The authors also acknowledge K. Nelson for critical reading of the manuscript and the many other important contributions in the drug discovery community that, regrettably, could not be cited owing to space constraints.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information S1 (table)
Selected examples of entities that promote translational research (PDF 113 kb)
Supplementary information S2 (table)
Common artefactual compound-mediated biological activities (PDF 118 kb)
Supplementary information S3 (table)
Assay types employed in drug discovery (PDF 81 kb)
Supplementary information S4 (table)
Examples of high-throughput screening assays (PDF 106 kb)
Glossary
- Chemical probe
-
A well-characterized compound showing context-related selectivity that potently modulates a biochemical target or pathway in cells or in vivo.
- Prodrugs
-
Compounds that are the metabolic precursors of drugs. Prodrugs can aid in drug delivery and can be new chemical entities.
- Biomarkers
-
Factors that are objectively measured and evaluated as indicators of biological or pathological processes, or of pharmacological responses to therapeutic intervention.
- Reporter gene assay
-
An assay in which an easily detectable reporter gene such as luciferase is fused to the promoter sequence of downstream target genes of the signalling pathway under investigation. Modulation of the pathway, such as activation or inhibition, will lead to changes in reporter gene expression (in the case of luciferase, these changes will be measured as luminescence).
- Orthogonal assays
-
Corresponding assays used following, or in parallel to, the primary high-throughput screening assay to confirm compound activity that is independent of the primary assay technique.
- Chemotypes
-
Chemical structure motifs or primary substructures that are common to a group of compounds.
- Diversity-oriented synthesis
-
Efficient synthesis of a collection (library) of structurally diverse and complex small molecules that differ in stereochemistry, functional groups and molecular framework.
- Pan-assay interference compounds
-
(PAINS). Compounds containing substructures that give rise to activity in assays irrespective of the biological target.
- Off-target activities
-
Activities of a compound that differ from its designed or annotated mechanism of action. Off-target activities are often undefined.
Rights and permissions
About this article
Cite this article
Dahlin, J., Inglese, J. & Walters, M. Mitigating risk in academic preclinical drug discovery. Nat Rev Drug Discov 14, 279–294 (2015). https://doi.org/10.1038/nrd4578
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrd4578
This article is cited by
-
Theoretical insight on antioxidant potency of kanzakiflavone-2 and its derivatives
Structural Chemistry (2021)
-
Early Detection of Toxic Cyanobacteria in Bulgarian Dam Water and In Vitro Evaluation of the Effect of Saponins From Astragalus glycyphyllos and A. glycyphylloides, in Cyanotoxin (Anatoxin-α)-Induced Neurotoxicity
Revista Brasileira de Farmacognosia (2020)
-
Computational advances in combating colloidal aggregation in drug discovery
Nature Chemistry (2019)
-
‘Stealth’ corporate innovation: an emerging threat for therapeutic drug development
Nature Immunology (2019)
-
Arylmethylamino steroids as antiparasitic agents
Nature Communications (2017)