Key Points
-
Prior efforts using genomics to inform drug discovery (that is, expressed sequencing tag profiling and genome-wide association studies) have yielded an array of potential targets but have encountered difficulty in translating these discoveries into clinically efficacious drugs.
-
Precision medicine marks a new relationship between genomics and drug discovery, one that provides both insights into the mechanisms and potential treatment options of a single patient's disease.
-
Oncology has been the leader in the field of precision medicine, with a multitude of successful examples of targeted treatments and immunotherapies available for a wide range of cancers.
-
Precision therapies for Mendelian diseases, such as those that replace deficient proteins, directly target the dysfunctional protein or disease-associated pathway or influence expression of disease-relevant genes, and are clinically available for a number of conditions, such as lysosomal storage disorders, cystic fibrosis, tuberous sclerosis and spinal muscular atrophy.
-
With various reliable model systems and a multitude of drug screening platforms, epilepsy is well positioned to serve as a model for precision medicine in highly genetic conditions.
-
Although it is currently uncertain how generalizable genomic precision medicine approaches will be for all diseases, we imagine that some therapeutics that were developed for defined genetic conditions will also be efficacious in individuals with mechanistically related disease that do not necessarily carry the same genetic drivers.
Abstract
For the past three decades, the use of genomics to inform drug discovery and development pipelines has generated both excitement and scepticism. Although earlier efforts successfully identified some new drug targets, the overall clinical efficacy of developed drugs has remained unimpressive, owing in large part to the heterogeneous causes of disease. Recent technological and analytical advances in genomics, however, have now made it possible to rapidly identify and interpret the genetic variation underlying a single patient's disease, thereby providing a window into patient-specific mechanisms that cause or contribute to disease, which could ultimately enable the 'precise' targeting of these mechanisms. Here, we first examine and highlight the successes and limitations of the earlier phases of genomics in drug discovery and development. We then review the current major efforts in precision medicine and discuss the potential broader utility of mechanistically guided treatments going forward.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
National Research Council (US) Committee on A Framework for Developing a New Taxonomy of Disease. Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease (National Academies Press, 2011).
Raviña Rubira, E. The Evolution of Drug Discovery: From Traditional Medicines to Modern Drugs. (Wiley-VCH, 2011).
Zanders, E. D. The Science and Business of Drug Discovery. (Springer, 2011).
Hopkins, M. M., Martin, P. A., Nightingale, P., Kraft, A. & Mahdi, S. The myth of the biotech revolution: an assessment of technological, clinical and organisational change. Res. Policy 36, 566–589 (2007).
Nightingale, P. & Madhi, S. in Knowledge Accumulation and Industry Evolution: The Case of Pharma-Biotech (eds Mazzucatu, M. & Dosi, G.) 73–111 (Cambridge Univ. Press, 2006).
Martin, P., Hopkins, M., Nightingale, P. & Kraft, A. in The Handbook of Genetics & Society: Mapping the New Genomic Era (eds Atkinson, P., Glasner, P. & Lock, M.) 145–162 (Routledge, 2009).
Nightingale, P. Economies of scale in experimentation: knowledge and technology in pharmaceutical R&D. Ind. Corp. Change 9, 315–359 (2000).
Debouck, C. & Metcalf, B. The impact of genomics on drug discovery. Annu. Rev. Pharmacol. Toxicol. 40, 193–207 (2000).
Adams, M. D. et al. Complementary DNA sequencing: expressed sequence tags and human genome project. Science 252, 1651–1656 (1991).
Adams, M. D. et al. Initial assessment of human gene diversity and expression patterns based upon 83 million nucleotides of cDNA sequence. Nature 377, 3–174 (1995).
Haseltine, W. A. Discovering genes for new medicines. Sci. Am. 276, 92–97 (1997).
Haseltine, W. A. Genomics and drug discovery. J. Am. Acad. Dermatol. 45, 473–475 (2001).
Gershon, D. Smithkline backs sequencing company. Nature 363, 387 (1993).
Ma, P. & Zemmel, R. Value of novelty? Nat. Rev. Drug Discov. 1, 571–572 (2002).
Hopkins, M., Kraft, A., Martin, P., Nightingale, P. & Madhi, S. in Comprehensive Medicinal Chemistry II (eds Taylor, J. B. & Triggle, D. J.) 591–613 (Elsevier, 2007).
Cook, D. et al. Lessons learned from the fate of AstraZeneca's drug pipeline: a five-dimensional framework. Nat. Rev. Drug Discov. 13, 419–431 (2014).
Nelson, M. R. et al. The support of human genetic evidence for approved drug indications. Nat. Genet. 47, 856–860 (2015).
Manolio, T. A., Brooks, L. D. & Collins, F. S. A. HapMap harvest of insights into the genetics of common disease. J. Clin. Invest. 118, 1590–1605 (2008).
Kingsmore, S. F., Lindquist, I. E., Mudge, J., Gessler, D. D. & Beavis, W. D. Genome-wide association studies: progress and potential for drug discovery and development. Nat. Rev. Drug Discov. 7, 221–230 (2008).
Lopes, M. C., Zeggini, E. & Panoutsopoulou, K. Do genome-wide association scans have potential for translation? Clin. Chem. Lab. Med. 50, 255–260 (2011).
Visscher, P. M., Brown, M. A., McCarthy, M. I. & Yang, J. Five years of GWAS discovery. Am. J. Hum. Genet. 90, 7–24 (2012).
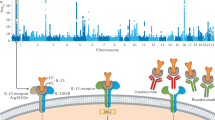
Duerr, R. H. et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science 314, 1461–1463 (2006).
Liu, Y. et al. A genome-wide association study of psoriasis and psoriatic arthritis identifies new disease loci. PLoS Genet. 4, e1000041 (2008).
Australo-Anglo-American Spondyloarthritis Consortium (TASC) et al. Genome-wide association study of ankylosing spondylitis identifies non-MHC susceptibility loci. Nat. Genet. 42, 123–127 (2010).
Gaffen, S. L., Jain, R., Garg, A. V. & Cua, D. J. The IL-23-IL-17 immune axis: from mechanisms to therapeutic testing. Nat. Rev. Immunol. 14, 585–600 (2014).
Feagan, B. G. et al. Ustekinumab as induction and maintenance therapy for Crohn's disease. N. Engl. J. Med. 375, 1946–1960 (2016).
Krueger, J. G. et al. Anti–IL-23A mAb BI 655066 for treatment of moderate-to-severe psoriasis: safety, efficacy, pharmacokinetics, and biomarker results of a single-rising-dose, randomized, double-blind, placebo-controlled trial. J. Allergy Clin. Immunol. 136, 116–124.e7 (2015).
Feagan, B. G. et al. Induction therapy with the selective interleukin-23 inhibitor risankizumab in patients with moderate-to-severe Crohn's disease: a randomised, double-blind, placebo-controlled phase 2 study. Lancet 389, 1699–1709 (2017).
Kopp, T. et al. Clinical improvement in psoriasis with specific targeting of interleukin-23. Nature 521, 222–226 (2015).
Poddubnyy, D., Hermann, K.-G. A., Callhoff, J., Listing, J. & Sieper, J. Ustekinumab for the treatment of patients with active ankylosing spondylitis: results of a 28-week, prospective, open-label, proof-of-concept study (TOPAS). Ann. Rheum. Dis. 73, 817–823 (2014).
Fragoulis, G. E., Siebert, S. & McInnes, I. B. Therapeutic targeting of IL-17 and IL-23 cytokines in immune-mediated diseases. Annu. Rev. Med. 67, 337–353 (2016).
Frayling, T. M. et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 316, 889–894 (2007).
Dina, C. et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat. Genet. 39, 724–726 (2007).
Scuteri, A. et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 3, e115 (2007).
Tung, Y. C. L., Yeo, G. S. H., O'Rahilly, S. & Coll, A. P. Obesity and FTO: changing focus at a complex locus. Cell Metab. 20, 710–718 (2014).
Loos, R. J. F. & Yeo, G. S. H. The bigger picture of FTO: the first GWAS-identified obesity gene. Nat. Rev. Endocrinol. 10, 51–61 (2014).
Claussnitzer, M. et al. FTO obesity variant circuitry and adipocyte browning in humans. N. Engl. J. Med. 373, 895–907 (2015).
Smemo, S. et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature 507, 371–375 (2014).
Boyle, E. A., Li, Y. I. & Pritchard, J. K. An expanded view of complex traits: from polygenic to omnigenic. Cell 169, 1177–1186 (2017). This paper proposes an 'omnigenic' model of complex disease and trait inheritance, arguing in favour of a few 'core' disease variants, with the bulk of heritability caused by an immense amount of 'peripheral' variants expressed in disease-relevant cell types.
Goldstein, D. B. Common genetic variation and human traits. N. Engl. J. Med. 360, 1696–1698 (2009).
Plenge, R. M., Scolnick, E. M. & Altshuler, D. Validating therapeutic targets through human genetics. Nat. Rev. Drug Discov. 12, 581–594 (2013).
Abifadel, M. et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat. Genet. 34, 154–156 (2003).
Fasano, T. et al. A novel loss of function mutation of PCSK9 gene in white subjects with low-plasma low-density lipoprotein cholesterol. Arterioscler. Thromb. Vasc. Biol. 27, 677–681 (2007).
Cohen, J. et al. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat. Genet. 37, 161–165 (2005).
Cohen, J. C., Boerwinkle, E., Mosley, T. H. & Hobbs, H. H. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N. Engl. J. Med. 354, 1264–1272 (2006).
Kotowski, I. K. et al. A spectrum of PCSK9 alleles contributes to plasma levels of low-density lipoprotein cholesterol. Am. J. Hum. Genet. 78, 410–422 (2006).
Zhang, D.-W. et al. Binding of proprotein convertase subtilisin/kexin type 9 to epidermal growth factor-like repeat A of low density lipoprotein receptor decreases receptor recycling and increases degradation. J. Biol. Chem. 282, 18602–18612 (2007).
Poirier, S. et al. The proprotein convertase PCSK9 induces the degradation of low density lipoprotein receptor (LDLR) and its closest family members VLDLR and ApoER2. J. Biol. Chem. 283, 2363–2372 (2008).
Dadu, R. T. & Ballantyne, C. M. Lipid lowering with PCSK9 inhibitors. Nat. Rev. Cardiol. 11, 563–575 (2014).
Sabatine, M. S. et al. Evolocumab and clinical outcomes in patients with cardiovascular disease. N. Engl. J. Med. 376, 1713–1722 (2017).
Kamb, A., Harper, S. & Stefansson, K. Human genetics as a foundation for innovative drug development. Nat. Biotechnol. 31, 975–978 (2013).
Eisenstein, M. Amgen and Regeneron push for a genetic renaissance in drug discovery. Nat. Biotechnol. 32, 208–209 (2014).
Sulem, P. et al. Identification of a large set of rare complete human knockouts. Nat. Genet. 47, 448–452 (2015).
Dewey, F. E. et al. Inactivating variants in ANGPTL4 and risk of coronary artery disease. N. Engl. J. Med. 374, 1123–1133 (2016).
Silver, R. T. et al. An evidence-based analysis of the effect of busulfan, hydroxyurea, interferon, and allogeneic bone marrow transplantation in treating the chronic phase of chronic myeloid leukemia: developed for the American Society of Hematology: presented in part at the Education Session of the American Society of Hematology, December 5, 1998, Miami Beach, FL. Blood 94, 1517–1536 (1999).
Stegmeier, F., Warmuth, M., Sellers, W. R. & Dorsch, M. Targeted cancer therapies in the twenty-first century: lessons from imatinib. Clin. Pharmacol. Ther. 87, 543–552 (2010).
Druker, B. J. Translation of the Philadelphia chromosome into therapy for CML. Blood 112, 4808–4817 (2008).
Demetri, G. D. et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N. Engl. J. Med. 347, 472–480 (2002).
Slamon, D. J. et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N. Engl. J. Med. 344, 783–792 (2001).
Hauschild, A. et al. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet 380, 358–365 (2012).
McArthur, G. A. et al. Safety and efficacy of vemurafenib in BRAF(V600E) and BRAF(V600K) mutation-positive melanoma (BRIM-3): extended follow-up of a phase 3, randomised, open-label study. Lancet Oncol. 15, 323–332 (2014).
Maemondo, M. et al. Gefitinib or chemotherapy for non–small-cell lung cancer with mutated EGFR. N. Engl. J. Med. 362, 2380–2388 (2010).
Zhou, C. et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 12, 735–742 (2011).
Mitsudomi, T. et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 11, 121–128 (2010).
Rosell, R. et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 13, 239–246 (2012).
Sequist, L. V. et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J. Clin. Oncol. 31, 3327–3334 (2013).
Wu, Y.-L. et al. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): an open-label, randomised phase 3 trial. Lancet Oncol. 15, 213–222 (2014).
Tewari, K. S., Eskander, R. N. & Monk, B. J. Development of olaparib for BRCA-deficient recurrent epithelial ovarian cancer. Clin. Cancer Res. 21, 3829–3835 (2015).
Lord, C. J. & Ashworth, A. PARP inhibitors: synthetic lethality in the clinic. Science 355, 1152–1158 (2017).
Pujade-Lauraine, E. et al. Treatment with olaparib monotherapy in the maintenance setting significantly improves progression-free survival in patients with platinum-sensitive relapsed ovarian cancer: results from the phase III SOLO2 study. Gynecol. Oncol. 145, 219–220 (2017).
Kaufman, B. et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J. Clin. Oncol. 33, 244–250 (2015).
Mirza, M. R. et al. Niraparib maintenance therapy in platinum-sensitive, recurrent ovarian cancer. N. Engl. J. Med. 375, 2154–2164 (2016).
Yver, A. Osimertinib (AZD9291) — a science-driven, collaborative approach to rapid drug design and development. Ann. Oncol. 27, 1165–1170 (2016).
Yang, Y. Cancer immunotherapy: harnessing the immune system to battle cancer. J. Clin. Invest. 125, 3335–3337 (2015).
Farkona, S., Diamandis, E. P. & Blasutig, I. M. Cancer immunotherapy: the beginning of the end of cancer? BMC Med. 14, 73 (2016).
Mullard, A. The cancer vaccine resurgence. Nat. Rev. Drug Discov. 15, 663–665 (2016).
van Rooij, N. et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J. Clin. Oncol. 31, e439–e442 (2013). This is the first report of T cell reactivity to tumour-specific neoantigens in human patients.
Snyder, A. et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 371, 2189–2199 (2014).
Linnemann, C. et al. High-throughput epitope discovery reveals frequent recognition of neo-antigens by CD4+ T cells in human melanoma. Nat. Med. 21, 81–85 (2014).
Gubin, M. M. et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature 515, 577–581 (2014).
Schumacher, T. N. & Schreiber, R. D. Neoantigens in cancer immunotherapy. Science 348, 69–74 (2015).
Rosenberg, S. A. et al. Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin. Cancer Res. 17, 4550–4557 (2011).
Dudley, M. E. et al. Adoptive cell transfer therapy following non-myeloablative but lymphodepleting chemotherapy for the treatment of patients with refractory metastatic melanoma. J. Clin. Oncol. 23, 2346–2357 (2005).
Dudley, M. E. et al. Cancer regression and autoimmunity in patients after clonal repopulation with antitumor lymphocytes. Science 298, 850–854 (2002).
Lu, Y.-C. et al. Mutated PPP1R3B is recognized by T cells used to treat a melanoma patient who experienced a durable complete tumor regression. J. Immunol. 190, 6034–6042 (2013).
Tran, E. et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science 344, 641–645 (2014).
Sadelain, M. CAR therapy: the CD19 paradigm. J. Clin. Invest. 125, 3392–3400 (2015).
Sharma, P. & Allison, J. P. Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell 161, 205–214 (2015).
Chen, D. S. & Mellman, I. Oncology meets immunology: the cancer-immunity cycle. Immunity 39, 1–10 (2013).
Brahmer, J. R. et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N. Engl. J. Med. 366, 2455–2465 (2012).
Powles, T. et al. MPDL3280A (anti-PD-L1) treatment leads to clinical activity in metastatic bladder cancer. Nature 515, 558–562 (2014).
Topalian, S. L. et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 366, 2443–2454 (2012).
Ansell, S. M. et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin's lymphoma. N. Engl. J. Med. 372, 311–319 (2015).
Seiwert, T. Y. et al. Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-012): an open-label, multicentre, phase 1b trial. Lancet Oncol. 17, 956–965 (2016).
Schadendorf, D. et al. Pooled analysis of long-term survival data from phase II and phase III trials of ipilimumab in unresectable or metastatic melanoma. J. Clin. Oncol. 33, 1889–1894 (2015).
Herbst, R. S. et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 387, 1540–1550 (2016).
Borghaei, H. et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N. Engl. J. Med. 373, 1627–1639 (2015).
Rebelatto, M. C. et al. Development of a programmed cell death ligand-1 immunohistochemical assay validated for analysis of non-small cell lung cancer and head and neck squamous cell carcinoma. Diagn. Pathol. 11, 95 (2016).
Massard, C. et al. Safety and efficacy of durvalumab (MEDI4736), an anti-programmed cell death ligand-1 immune checkpoint inhibitor, in patients with advanced urothelial bladder cancer. J. Clin. Oncol. 34, 3119–3125 (2016).
Rosenberg, J. E. et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet 387, 1909–1920 (2016).
Vanneman, M. & Dranoff, G. Combining immunotherapy and targeted therapies in cancer treatment. Nat. Rev. Cancer 12, 237–251 (2012). This is an insightful review that highlights the opportunities for and potential clinical benefits of combined targeted therapies and immunotherapies in oncology precision medicine.
Weber, J. S. et al. Nivolumab versus chemotherapy in patients with advanced melanoma who progressed after anti-CTLA-4 treatment (CheckMate 037): a randomised, controlled, open-label, phase 3 trial. Lancet Oncol. 16, 375–384 (2015).
Hammers, H. J. et al. Phase I study of nivolumab in combination with ipilimumab in metastatic renal cell carcinoma (mRCC). J. Clin. Oncol. 32, 4504–4504 (2014).
Rakhra, K. et al. CD4+ T cells contribute to the remodeling of the microenvironment required for sustained tumor regression upon oncogene inactivation. Cancer Cell 18, 485–498 (2010).
Hughes, P. E., Caenepeel, S. & Wu, L. C. Targeted therapy and checkpoint immunotherapy combinations for the treatment of cancer. Trends Immunol. 37, 462–476 (2016).
Renfro, L. A. & Sargent, D. J. Statistical controversies in clinical research: basket trials, umbrella trials, and other master protocols: a review and examples. Ann. Oncol. 28, 34–43 (2017).
Schork, N. J. Personalized medicine: time for one-person trials. Nature 520, 609–611 (2015).
Rubin, R. A. Precision medicine approach to clinical trials. JAMA 316, 1953–1955 (2016).
Alvarez, M. J. et al. Functional characterization of somatic mutations in cancer using network-based inference of protein activity. Nat. Genet. 48, 838–847 (2016).
Kornfeld, S. Lysosomal enzyme targeting. Biochem. Soc. Trans. 18, 367–374 (1990).
Desnick, R. J. & Schuchman, E. H. Enzyme replacement and enhancement therapies: lessons from lysosomal disorders. Nat. Rev. Genet. 3, 954–966 (2002).
Van Goor, F. et al. Rescue of CF airway epithelial cell function in vitro by a CFTR potentiator, VX-770. Proc. Natl Acad. Sci. USA 106, 18825–18830 (2009). This study reports the in vitro mechanism of the now-approved cystic fibrosis drug ivacaftor (VX-770) that selectively targets the molecular defect underlying the G551D mutation in CFTR.
Germain, D. P. et al. Treatment of Fabry's disease with the pharmacologic chaperone migalastat. N. Engl. J. Med. 375, 545–555 (2016).
Hughes, D. A. et al. Oral pharmacological chaperone migalastat compared with enzyme replacement therapy in Fabry disease: 18-month results from the randomised phase III ATTRACT study. J. Med. Genet. 54, 288–296 (2016).
French, J. A. et al. Adjunctive everolimus therapy for treatment-resistant focal-onset seizures associated with tuberous sclerosis (EXIST-3): a phase 3, randomised, double-blind, placebo-controlled study. Lancet 388, 2153–2163 (2016). This study reports clinical trial data for the use of the mTOR inhibitor everolimus in the treatment of TSC-related seizures, thus serving as an example of pathway-based treatment.
Hua, Y., Vickers, T. A., Okunola, H. L., Bennett, C. F. & Krainer, A. R. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am. J. Hum. Genet. 82, 834–848 (2008). This study describes the use of an ASO tiling method to interfere with the splicing silencers heterogeneous nuclear ribonucleoprotein A1 (HNRNPA1) and HNRNPA2 as a therapeutic mechanism for SMA and provides an example of how drugs may be designed to exert effects at the level of gene expression.
Corey, D. R. Nusinersen, an antisense oligonucleotide drug for spinal muscular atrophy. Nat. Neurosci. 20, 497–499 (2017).
Brodie, M. J., Barry, S. J. E., Bamagous, G. A., Norrie, J. D. & Kwan, P. Patterns of treatment response in newly diagnosed epilepsy. Neurology 78, 1548–1554 (2012).
Sankaraneni, R. & Lachhwani, D. Antiepileptic drugs — a review. Pediatr. Ann. 44, e36–e42 (2015).
Epi4K Consortium et al. De novo mutations in epileptic encephalopathies. Nature 501, 217–221 (2013).
Epilepsy Phenome/Genome Project, Epi4K Consortium & EuroEPINOMICS-RES Consortium. De novo mutations in synaptic transmission genes including DNM1 cause epileptic encephalopathies. Am. J. Hum. Genet. 100, 179 (2017).
EpiPM Consortium. A roadmap for precision medicine in the epilepsies. Lancet Neurol. 14, 1219–1228 (2015). This article provides a conceptual framework for developing and implementing targeted therapies for epilepsy.
Dhindsa, R. S. & Goldstein, D. B. Genetic discoveries drive molecular analyses and targeted therapeutic options in the epilepsies. Curr. Neurol. Neurosci. Rep. 15, 70 (2015).
Yang, B. et al. Pharmacological activation and inhibition of Slack (Slo2.2) channels. Neuropharmacology 51, 896–906 (2006).
Milligan, C. J. et al. KCNT1 gain of function in 2 epilepsy phenotypes is reversed by quinidine. Ann. Neurol. 75, 581–590 (2014).
Bearden, D. et al. Targeted treatment of migrating partial seizures of infancy with quinidine. Ann. Neurol. 76, 457–461 (2014).
Mikati, M. A. et al. Quinidine in the treatment of KCNT1-positive epilepsies. Ann. Neurol. 78, 995–999 (2015).
Pierson, T. M. et al. GRIN2A mutation and early-onset epileptic encephalopathy: personalized therapy with memantine. Ann. Clin. Transl Neurol. 1, 190–198 (2014).
Li, D. et al. GRIN2D recurrent de novo dominant mutation causes a severe epileptic encephalopathy treatable with NMDA receptor channel blockers. Am. J. Hum. Genet. 99, 802–816 (2016).
Wang, H. S. et al. KCNQ2 and KCNQ3 potassium channel subunits: molecular correlates of the M-channel. Science 282, 1890–1893 (1998).
Orhan, G. et al. Dominant-negative effects of KCNQ2 mutations are associated with epileptic encephalopathy. Ann. Neurol. 75, 382–394 (2014).
Ihara, Y. et al. Retigabine, a Kv7.2/Kv7.3-channel opener, attenuates drug-induced seizures in knock-in mice harboring Kcnq2 mutations. PLoS ONE 11, e0150095 (2016).
Wuttke, T. V. et al. Peripheral nerve hyperexcitability due to dominant-negative KCNQ2 mutations. Neurology 69, 2045–2053 (2007).
Millichap, J. J. et al. KCNQ2 encephalopathy: features, mutational hot spots, and ezogabine treatment of 11 patients. Neurol. Genet. 2, e96 (2016).
International League Against Epilepsy Consortium on Complex Epilepsies. Genetic determinants of common epilepsies: a meta-analysis of genome-wide association studies. Lancet Neurol. 13, 893–903 (2014). This meta-analysis implicates mutations in genes initially known to cause the severe, monogenic epilepsies in the more common generalized and focal epilepsies, thus serving as an example of the allelic and corresponding phenotypic heterogeneity that may render certain targeted treatments applicable to patients carrying a range of mutations in the same gene.
Iossifov, I. et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature 515, 216–221 (2014).
De Rubeis, S. et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515, 209–215 (2014).
Zaidi, S. et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature 498, 220–223 (2013).
Al Turki, S. et al. Rare variants in NR2F2 cause congenital heart defects in humans. Am. J. Hum. Genet. 94, 574–585 (2014).
Petrovski, S. et al. An exome sequencing study to assess the role of rare genetic variation in pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 196, 82–93 (2017).
Asgari, S. et al. Exome sequencing reveals primary immunodeficiencies in children with community-acquired Pseudomonas aeruginosa sepsis. Front. Immunol. 7, 357 (2016).
Maffucci, P. et al. Genetic diagnosis using whole exome sequencing in common variable immunodeficiency. Front. Immunol. 7, 220 (2016).
Dickinson, R. E. et al. Exome sequencing identifies GATA-2 mutation as the cause of dendritic cell, monocyte, B and NK lymphoid deficiency. Blood 118, 2656–2658 (2011).
Marouli, E. et al. Rare and low-frequency coding variants alter human adult height. Nature 542, 186–190 (2017).
Surakka, I. et al. The impact of low-frequency and rare variants on lipid levels. Nat. Genet. 47, 589–597 (2015).
Surendran, P. et al. Trans-ancestry meta-analyses identify rare and common variants associated with blood pressure and hypertension. Nat. Genet. 48, 1151–1161 (2016).
Weishaupt, J. H., Hyman, T. & Dikic, I. Common molecular pathways in amyotrophic lateral sclerosis and frontotemporal dementia. Trends Mol. Med. 22, 769–783 (2016). This review summarizes findings from amyotrophic lateral sclerosis genetic studies and the convergence of implicated genes into functional pathways, thus underscoring the potential utility of these disease-associated pathways as therapeutic targets.
Oza, A. M. et al. Olaparib combined with chemotherapy for recurrent platinum-sensitive ovarian cancer: a randomised phase 2 trial. Lancet Oncol. 16, 87–97 (2015).
Brunkow, M. E. et al. Bone dysplasia sclerosteosis results from loss of the SOST gene product, a novel cystine knot-containing protein. Am. J. Hum. Genet. 68, 577–589 (2001).
Winkler, D. G. et al. Osteocyte control of bone formation via sclerostin, a novel BMP antagonist. EMBO J. 22, 6267–6276 (2003).
Padhi, D., Jang, G., Stouch, B., Fang, L. & Posvar, E. Single-dose, placebo-controlled, randomized study of AMG 785, a sclerostin monoclonal antibody. J. Bone Miner. Res. 26, 19–26 (2011).
McClung, M. R. et al. Romosozumab in postmenopausal women with low bone mineral density. N. Engl. J. Med. 370, 412–420 (2014).
Cosman, F. et al. Romosozumab treatment in postmenopausal women with osteoporosis. N. Engl. J. Med. 375, 1532–1543 (2016).
Yang, Y. et al. Mutations in SCN9A, encoding a sodium channel alpha subunit, in patients with primary erythermalgia. J. Med. Genet. 41, 171–174 (2004).
Fertleman, C. R. et al. SCN9A mutations in paroxysmal extreme pain disorder: allelic variants underlie distinct channel defects and phenotypes. Neuron 52, 767–774 (2006).
Cox, J. J. et al. An SCN9A channelopathy causes congenital inability to experience pain. Nature 444, 894–898 (2006).
Reimann, F. et al. Pain perception is altered by a nucleotide polymorphism in SCN9A. Proc. Natl Acad. Sci. USA 107, 5148–5153 (2010).
Duan, G. et al. A single-nucleotide polymorphism in SCN9A may decrease postoperative pain sensitivity in the general population. Anesthesiology 118, 436–442 (2013).
Acknowledgements
The authors thank J. Carulli, M. Lalioti and C. Bostick for their valuable feedback on this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
D.G. is a founder of and holds equity in Pairnomix and Praxis, serves as a consultant to AstraZeneca and has received research support from Janssen, Gilead, Biogen, AstraZeneca and UCB. A.P. is a paid employee of and holds stock in AstraZeneca.
Related links
FURTHER INFORMATION
Rights and permissions
About this article
Cite this article
Dugger, S., Platt, A. & Goldstein, D. Drug development in the era of precision medicine. Nat Rev Drug Discov 17, 183–196 (2018). https://doi.org/10.1038/nrd.2017.226
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrd.2017.226