Key Points
-
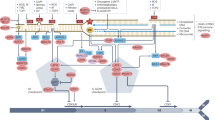
Several cancer chemotherapy drugs work by producing excessive DNA damage that causes cell death directly or following DNA replication. Survival is promoted through repair of these lesions by a number of DNA repair pathways.
-
The efficacy of anticancer drugs is highly influenced by cellular DNA repair capacity. Inhibitors of DNA repair increase the efficacy of DNA-damaging anticancer drugs in preclinical models. Small-molecule inhibitors of DNA repair have been combined with conventional chemotherapy drugs in several phase I–II clinical trials.
-
Tumour development can be associated with perturbed DNA damage response and repair pathways. This perturbation results in reduced DNA repair capacity and increased genetic instability in tumour cells. Defects in one DNA repair pathway can be compensated for by other pathways. Such compensating pathways can be identified in synthetic lethality screens and then specifically targeted for treatment of DNA repair-defective tumours.
-
Evidence indicates that inhibitors of DNA repair pathways can work as single agents for the targeted treatment of DNA repair-defective cancers. This hypothesis is currently being tested in phase II trials in which patients with breast or ovarian cancers that are defective in homologous recombination are being treated with a poly(ADP-ribose) polymerase inhibitor.
-
Tumours often exhibit replication stress as a consequence of oncogene-induced growth signals or hypoxia-induced replication arrest. We propose that DNA repair inhibitors could be used to prevent the repair of replication lesions present in tumour cells and convert them into fatal replication lesions that specifically kill cancer cells.
Abstract
DNA repair pathways can enable tumour cells to survive DNA damage that is induced by chemotherapeutic treatments; therefore, inhibitors of specific DNA repair pathways might prove efficacious when used in combination with DNA-damaging chemotherapeutic drugs. In addition, alterations in DNA repair pathways that arise during tumour development can make some cancer cells reliant on a reduced set of DNA repair pathways for survival. There is evidence that drugs that inhibit one of these pathways in such tumours could prove useful as single-agent therapies, with the potential advantage that this approach could be selective for tumour cells and have fewer side effects.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Hsiang, Y. H., Lihou, M. G. & Liu, L. F. Arrest of replication forks by drug-stabilized topoisomerase I — DNA cleavable complexes as a mechanism of cell killing by camptothecin. Cancer Res. 49, 5077–5082 (1989).
Markovits, J. et al. Topoisomerase II-mediated DNA breaks and cytotoxicity in relation to cell proliferation and the cell cycle in NIH 3T3 fibroblasts and L1210 leukemia cells. Cancer Res. 47, 2050–2055 (1987).
Ikegami, S. et al. Aphidicolin prevents mitotic cell division by interfering with the activity of DNA polymerase-α. Nature 275, 458–460 (1978).
Bianchi, V., Pontis, E. & Reichard, P. Changes of deoxyribonucleoside triphosphate pools induced by hydroxyurea and their relation to DNA synthesis. J. Biol. Chem. 261, 16037–16042 (1986).
Lundin, C. et al. Different roles for nonhomologous end joining and homologous recombination following replication arrest in mammalian cells. Mol. Cell Biol. 22, 5869–5878 (2002).
Saintigny, Y. et al. Characterization of homologous recombination induced by replication inhibition in mammalian cells. EMBO J. 20, 3861–3870 (2001).
Swann, P. F. et al. Role of postreplicative DNA mismatch repair in the cytotoxic action of thioguanine. Science 273, 1109–1111 (1996).
Wang, J. C. Cellular roles of DNA topoisomerases: a molecular perspective. Nature Rev. Mol. Cell Biol. 3, 430–440 (2002).
Painter, R. B. & Cleaver, J. E. Repair replication in HeLa cells after large doses of x-irradiation. Nature 216, 369–370 (1967).
Canman, C. E. et al. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 281, 1677–1679 (1998).
Falck, J., Mailand, N., Syljuasen, R. G., Bartek, J. & Lukas, J. The ATM–Chk2–Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature 410, 842–847 (2001). This paper describes the molecular mechanism by which ATM rapidly inactivates DNA synthesis following ionizing radiation.
Cliby, W. A. et al. Overexpression of a kinase-inactive ATR protein causes sensitivity to DNA-damaging agents and defects in cell cycle checkpoints. EMBO J. 17, 159–169 (1998).
Taylor, A. M. et al. Ataxia telangiectasia: a human mutation with abnormal radiation sensitivity. Nature 258, 427–429 (1975).
Sargent, R. G., Brenneman, M. A. & Wilson, J. H. Repair of site-specific double-strand breaks in a mammalian chromosome by homologous and illegitimate recombination. Mol. Cell Biol. 17, 267–277 (1997).
Arnaudeau, C., Lundin, C. & Helleday, T. DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. J. Mol. Biol. 307, 1235–1245 (2001).
Sharma, R. A. & Dianov, G. L. Targeting base excision repair to improve cancer therapies. Mol. Aspects Med. 28, 345–374 (2007).
Huang, J. C., Svoboda, D. L., Reardon, J. T. & Sancar, A. Human nucleotide excision nuclease removes thymine dimers from DNA by incising the 22nd phosphodiester bond 5′ and the 6th phosphodiester bond 3′ to the photodimer. Proc. Natl Acad. Sci. USA 89, 3664–3668 (1992).
Sugasawa, K. et al. A multistep damage recognition mechanism for global genomic nucleotide excision repair. Genes Dev. 15, 507–521 (2001).
Sedgwick, B. Repairing DNA-methylation damage. Nature Rev. Mol. Cell Biol. 5, 148–157 (2004).
Lindahl, T., Demple, B. & Robins, P. Suicide inactivation of the E. coli O6-methylguanine-DNA methyltransferase. EMBO J. 1, 1359–1363 (1982).
Duncan, T. et al. Reversal of DNA alkylation damage by two human dioxygenases. Proc. Natl Acad. Sci. USA 99, 16660–16665 (2002).
Karran, P. & Marinus, M. G. Mismatch correction at O6-methylguanine residues in E. coli DNA. Nature 296, 868–869 (1982).
Yoshioka, K., Yoshioka, Y. & Hsieh, P. ATR kinase activation mediated by MutSα and MutLα in response to cytotoxic O6-methylguanine adducts. Mol. Cell 22, 501–510 (2006).
Fram, R. J., Cusick, P. S., Wilson, J. M. & Marinus, M. G. Mismatch repair of cis-diamminedichloroplatinum(II)-induced DNA damage. Mol. Pharmacol. 28, 51–55 (1985).
Masutani, C., Kusumoto, R., Iwai, S. & Hanaoka, F. Mechanisms of accurate translesion synthesis by human DNA polymerase η. EMBO J. 19, 3100–3109 (2000).
Vaisman, A., Masutani, C., Hanaoka, F. & Chaney, S. G. Efficient translesion replication past oxaliplatin and cisplatin GpG adducts by human DNA polymerase η. Biochemistry 39, 4575–4580 (2000).
Fukui, T. et al. Distinct roles of DNA polymerases δ and ɛ at the replication fork in Xenopus egg extracts. Genes Cells 9, 179–191 (2004).
Pursell, Z. F., Isoz, I., Lundstrom, E. B., Johansson, E. & Kunkel, T. A. Yeast DNA polymerase ɛ participates in leading-strand DNA replication. Science 317, 127–130 (2007).
Lehmann, A. R. Translesion synthesis in mammalian cells. Exp. Cell Res. 312, 2673–2676 (2006).
Sorensen, C. S. et al. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nature Cell Biol. 7, 195–201 (2005).
Kastan, M. B. & Bartek, J. Cell-cycle checkpoints and cancer. Nature 432, 316–323 (2004).
Bartkova, J. et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 444, 633–637 (2006).
Di Micco, R. et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 444, 638–642 (2006). References 32 and 33 show that oncogenes induce replication lesions in premalignant cancer cells that in turn activate senescence as a tumour barrier.
Arnaudeau, C., Tenorio Miranda, E., Jenssen, D. & Helleday, T. Inhibition of DNA synthesis is a potent mechanism by which cytostatic drugs induce homologous recombination in mammalian cells. Mutat. Res. DNA Repair 461, 221–228 (2000).
Helleday, T., Lo, J., van Gent, D. C. & Engelward, B. P. DNA double-strand break repair: From mechanistic understanding to cancer treatment. DNA Repair (Amst.) 6, 923–935 (2007).
Patel, K. J. & Joenje, H. Fanconi anemia and DNA replication repair. DNA Repair (Amst.) 6, 885–890 (2007).
Hanada, K. et al. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nature Struct. Mol. Biol. 14, 1096–1104 (2007).
Karow, J. K., Constantinou, A., Li, J. L., West, S. C. & Hickson, I. D. The Bloom's syndrome gene product promotes branch migration of holliday junctions. Proc. Natl Acad. Sci. USA 97, 6504–6508 (2000).
Lebel, M., Spillare, E. A., Harris, C. C. & Leder, P. The Werner syndrome gene product co-purifies with the DNA replication complex and interacts with PCNA and topoisomerase I. J. Biol. Chem. 274, 37795–37799 (1999).
Constantinou, A. et al. Werner's syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep. 1, 80–84 (2000).
Wu, L. & Hickson, I. D. DNA helicases required for homologous recombination and repair of damaged replication forks. Annu. Rev. Genet. 40, 279–306 (2006).
Niedzwiedz, W. et al. The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Mol. Cell 15, 607–620 (2004).
Wu, L. & Hickson, I. D. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature 426, 870–874 (2003).
Chen, X. B. et al. Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol. Cell 8, 1117–1127 (2001).
Hinz, J. M., Nham, P. B., Urbin, S. S., Jones, I. M. & Thompson, L. H. Disparate contributions of the Fanconi anemia pathway and homologous recombination in preventing spontaneous mutagenesis. Nucleic Acids Res. 35, 3733–3740 (2007).
Thompson, L. H. Strategies for cloning mammalian DNA repair genes. Methods Mol. Biol. 113, 57–85 (1999).
Chabner, B. A. & Roberts, T. G. Jr. Chemotherapy and the war on cancer. Nature Rev. Cancer 5, 65–72 (2005).
Stevens, M. F. et al. Antitumor activity and pharmacokinetics in mice of 8-carbamoyl-3-methyl-imidazo[5,1-d]-1,2,3,5-tetrazin-4(3H)-one (CCRG 81045; M & B 39831), a novel drug with potential as an alternative to dacarbazine. Cancer Res. 47, 5846–5852 (1987).
Stupp, R. et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 352, 987–996 (2005). References 48 and 49 demonstrate the timescale required from drug discovery in the laboratory to the demonstration of improved survival in patients in a randomized phase III clinical trial, in this case for temozolomide as a radiosensitizer.
Dolan, M. E. & Pegg, A. E. O6-Benzylguanine and its role in chemotherapy. Clin. Cancer Res. 3, 837–847 (1997).
Gerson, S. L., Berger, N. A., Arce, C., Petzold, S. J. & Willson, J. K. Modulation of nitrosourea resistance in human colon cancer by O6-methylguanine. Biochem. Pharmacol. 43, 1101–1107 (1992).
Middleton, M. R. & Margison, G. P. Improvement of chemotherapy efficacy by inactivation of a DNA-repair pathway. Lancet Oncol. 4, 37–44 (2003).
Ranson, M. et al. Lomeguatrib, a potent inhibitor of O6-alkylguanine-DNA-alkyltransferase: phase I safety, pharmacodynamic, and pharmacokinetic trial and evaluation in combination with temozolomide in patients with advanced solid tumors. Clin. Cancer Res. 12, 1577–1584 (2006).
Quinn, J. A. et al. Phase I trial of temozolomide plus O6-benzylguanine for patients with recurrent or progressive malignant glioma. J. Clin. Oncol. 23, 7178–7187 (2005).
Khan, O. & Middleton, M. R. The therapeutic potential of O6-alkylguanine DNA alkyltransferase inhibitors. Expert Opin. Investig. Drugs 16, 1573–1584 (2007).
Durkacz, B. W., Omidiji, O., Gray, D. A. & Shall, S. (ADP-ribose)n participates in DNA excision repair. Nature 283, 593–596 (1980). This study was first to demonstrate that PARP inhibitors can be used to increase the toxicity of DNA-damaging agents.
Yang, Y. G., Cortes, U., Patnaik, S., Jasin, M. & Wang, Z. Q. Ablation of PARP-1 does not interfere with the repair of DNA double-strand breaks, but compromises the reactivation of stalled replication forks. Oncogene 23, 3872–3882 (2004).
Ahel, I. et al. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature 451, 81–85 (2008).
Rosenberg, B., VanCamp, L., Trosko, J. E. & Mansour, V. H. Platinum compounds: a new class of potent antitumour agents. Nature 222, 385–386 (1969).
Kubo, S. et al. Participation of poly(ADP-ribose) polymerase in the drug sensitivity in human lung cancer cell lines. J. Cancer Res. Clin. Oncol. 118, 244–248 (1992).
Miknyoczki, S. J. et al. Chemopotentiation of temozolomide, irinotecan, and cisplatin activity by CEP-6800, a poly(ADP-ribose) polymerase inhibitor. Mol. Cancer Ther. 2, 371–382 (2003).
Robins, H. I. et al. Phase I trial of intravenous thymidine and carboplatin in patients with advanced cancer. J. Clin. Oncol. 17, 2922–2931 (1999).
Donawho, C. K. et al. ABT-888, an orally active poly(ADP-ribose) polymerase inhibitor that potentiates DNA-damaging agents in preclinical tumor models. Clin. Cancer Res. 13, 2728–2737 (2007).
Gifford, G., Paul, J., Vasey, P. A., Kaye, S. B. & Brown, R. The acquisition of hMLH1 methylation in plasma DNA after chemotherapy predicts poor survival for ovarian cancer patients. Clin. Cancer Res. 10, 4420–4426 (2004).
Plumb, J. A., Strathdee, G., Sludden, J., Kaye, S. B. & Brown, R. Reversal of drug resistance in human tumor xenografts by 2′-deoxy-5-azacytidine-induced demethylation of the hMLH1 gene promoter. Cancer Res. 60, 6039–6044 (2000).
Rabik, C. A. & Dolan, M. E. Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat. Rev. 33, 9–23 (2007).
Olaussen, K. A. et al. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N. Engl. J. Med. 355, 983–991 (2006).
Tsao, M. S. et al. Prognostic and predictive importance of p53 and RAS for adjuvant chemotherapy in non small-cell lung cancer. J. Clin. Oncol. 25, 5240–5247 (2007).
Jiang, H. & Yang, L. Y. Cell cycle checkpoint abrogator UCN-01 inhibits DNA repair: association with attenuation of the interaction of XPA and ERCC1 nucleotide excision repair proteins. Cancer Res. 59, 4529–4534 (1999).
Matthews, D. J. et al. Pharmacological abrogation of S-phase checkpoint enhances the anti-tumor activity of gemcitabine in vivo. Cell Cycle 6, 104–110 (2007).
Syljuasen, R. G. et al. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol. Cell Biol. 25, 3553–3562 (2005).
Hartley, K. O. et al. DNA-dependent protein kinase catalytic subunit: a relative of phosphatidylinositol 3-kinase and the ataxia telangiectasia gene product. Cell 82, 849–856 (1995).
Blunt, T. et al. Defective DNA-dependent protein kinase activity is linked to VDJ recombination and DNA repair defects associated with the murine scid mutation. Cell 80, 813–823 (1995). This paper identifies DNAPK and shows that it can be used as a target to increase toxicity following ionizing radiation.
Monfar, M. et al. Activation of pp70/85 S6 kinases in interleukin-2-responsive lymphoid cells is mediated by phosphatidylinositol 3-kinase and inhibited by cyclic AMP. Mol. Cell Biol. 15, 326–337 (1995).
Wipf, P. & Halter, R. J. Chemistry and biology of wortmannin. Org. Biomol. Chem. 3, 2053–2061 (2005).
Leahy, J. J. et al. Identification of a highly potent and selective DNA-dependent protein kinase (DNA-PK) inhibitor (NU7441) by screening of chromenone libraries. Bioorg. Med. Chem. Lett. 14, 6083–6087 (2004).
Zhao, Y. et al. Preclinical evaluation of a potent novel DNA-dependent protein kinase inhibitor NU7441. Cancer Res. 66, 5354–5362 (2006).
Farmer, H. et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 (2005).
Bryant, H. E. et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose)polymerase. Nature 434, 913–917 (2005). References 78 and 79 show that PARP inhibitors can be used to selectively kill BRCA1- or BRCA2-defective tumours.
Moynahan, M. E., Chiu, J. W., Koller, B. H. & Jasin, M. Brca1 controls homology-directed DNA repair. Mol. Cell 4, 511–518 (1999).
Moynahan, M. E., Pierce, A. J. & Jasin, M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell 7, 263–272 (2001).
Patel, K. J. et al. Involvement of Brca2 in DNA repair. Mol. Cell 1, 347–357 (1998).
Lomonosov, M., Anand, S., Sangrithi, M., Davies, R. & Venkitaraman, A. R. Stabilization of stalled DNA replication forks by the BRCA2 breast cancer susceptibility protein. Genes Dev. 17, 3017–3022 (2003).
Helleday, T., Bryant, H. E. & Schultz, N. Poly(ADP-ribose) polymerase (PARP-1) in homologous recombination and as a target for cancer therapy. Cell Cycle 4, 1176–1178 (2005).
Schultz, N., Lopez, E., Saleh-Gohari, N. & Helleday, T. Poly(ADP-ribose) polymerase (PARP-1) has a controlling role in homologous recombination. Nucleic Acids Res. 31, 4959–4964 (2003).
Fisher, A., Hochegger, H., Takeda, S. & Caldecott, K. W. Poly (ADP-ribose) polymerase-1 accelerates single-strand break repair in concert with poly (ADP-ribose) glycohydrolase. Mol. Cell Biol. 27, 5597–5605 (2007).
McCabe, N. et al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 66, 8109–8115 (2006).
Bryant, H. E. & Helleday, T. Inhibition of poly (ADP-ribose) polymerase activates ATM which is required for subsequent homologous recombination repair. Nucleic Acids Res. 34, 1685–1691 (2006).
Kennedy, R. D. et al. Fanconi anemia pathway-deficient tumor cells are hypersensitive to inhibition of ataxia telangiectasia mutated. J. Clin. Invest. 117, 1440–1449 (2007).
Lengauer, C., Kinzler, K. W. & Vogelstein, B. Genetic instabilities in human cancers. Nature 396, 643–649 (1998).
Gorgoulis, V. G. et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 434, 907–913 (2005).
Bartkova, J. et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434, 864–870 (2005).
Kinzler, K. W. & Vogelstein, B. Cancer-susceptibility genes. Gatekeepers and caretakers. Nature 386, 761–763 (1997).
Schmitt, C. A. Senescence, apoptosis and therapy — cutting the lifelines of cancer. Nature Rev. Cancer 3, 286–295 (2003).
Braig, M. et al. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature 436, 660–665 (2005).
Vousden, K. H. & Lane, D. P. p53 in health and disease. Nature Rev. Mol. Cell Biol. 8, 275–283 (2007).
Mariani, B. D. & Schimke, R. T. Gene amplification in a single cell cycle in Chinese hamster ovary cells. J. Biol. Chem. 259, 1901–1910 (1984).
Rambach, W. A., Cooper, J. A. & Alt, H. L. Effect of hypoxia on DNA synthesis in the bone marrow and spleen of the rat. Science 119, 380–381 (1954).
Hammond, E. M., Denko, N. C., Dorie, M. J., Abraham, R. T. & Giaccia, A. J. Hypoxia links ATR and p53 through replication arrest. Mol. Cell Biol. 22, 1834–1843 (2002).
Hammond, E. M., Dorie, M. J. & Giaccia, A. J. ATR/ATM targets are phosphorylated by ATR in response to hypoxia and ATM in response to reoxygenation. J. Biol. Chem. 278, 12207–12213 (2003).
Gibson, S. L., Bindra, R. S. & Glazer, P. M. Hypoxia-induced phosphorylation of Chk2 in an ataxia telangiectasia mutated-dependent manner. Cancer Res. 65, 10734–10741 (2005).
Bindra, R. S., Crosby, M. E. & Glazer, P. M. Regulation of DNA repair in hypoxic cancer cells. Cancer Metastasis Rev. 26, 249–260 (2007).
Rockwell, S., Yuan, J., Peretz, S. & Glazer, P. M. Genomic instability in cancer. Novartis Found. Symp. 240, 133–142; discussion 142–151 (2001).
Reynolds, T. Y., Rockwell, S. & Glazer, P. M. Genetic instability induced by the tumor microenvironment. Cancer Res. 56, 5754–5757 (1996).
Hammond, E. M., Dorie, M. J. & Giaccia, A. J. Inhibition of ATR leads to increased sensitivity to hypoxia/reoxygenation. Cancer Res. 64, 6556–6562 (2004).
Hanahan, D. & Weinberg, R. A. The hallmarks of cancer. Cell 100, 57–70 (2000).
Di Micco, R., Fumagalli, M. & di Fagagna, F. Breaking news: high-speed race ends in arrest — how oncogenes induce senescence. Trends Cell Biol. 17, 529–536 (2007).
Giannini, G. et al. Human MRE11 is inactivated in mismatch repair-deficient cancers. EMBO Rep. 3, 248–254 (2002).
Shor, E. et al. Mutations in homologous recombination genes rescue top3 slow growth in Saccharomyces cerevisiae. Genetics 162, 647–662 (2002).
Wallis, J. W., Chrebet, G., Brodsky, G., Rolfe, M. & Rothstein, R. A hyper-recombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell 58, 409–419 (1989).
Tong, A. H. et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294, 2364–2368 (2001). This study show how systematic genetic analysis can be performed to produce a global map of gene function.
Miki, Y. et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266, 66–71 (1994).
Hiramoto, T. et al. Mutations of a novel human RAD54 homologue, RAD54B, in primary cancer. Oncogene 18, 3422–3426 (1999).
Schoenmakers, E. F., Huysmans, C. & Van de Ven, W. J. Allelic knockout of novel splice variants of human recombination repair gene RAD51B in t(12;14) uterine leiomyomas. Cancer Res. 59, 19–23 (1999).
Wong, A. K. et al. Characterization of a carboxy-terminal BRCA1 interacting protein. Oncogene 17, 2279–2285 (1998).
Riballo, E. et al. Identification of a defect in DNA ligase IV in a radiosensitive leukaemia patient. Curr. Biol. 9, 699–702 (1999).
Moshous, D. et al. Partial T and B lymphocyte immunodeficiency and predisposition to lymphoma in patients with hypomorphic mutations in Artemis. J. Clin. Invest. 111, 381–387 (2003).
Nicolaides, N. C. et al. Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature 371, 75–80 (1994).
Lipkin, S. M. et al. Germline and somatic mutation analyses in the DNA mismatch repair gene MLH3: Evidence for somatic mutation in colorectal cancers. Hum. Mutat. 17, 389–396 (2001).
de la Chapelle, A. Genetic predisposition to colorectal cancer. Nature Rev. Cancer 4, 769–780 (2004).
German, J., Bloom, D. & Passarge, E. Bloom's syndrome. V. Surveillance for cancer in affected families. Clin. Genet. 12, 162–168 (1977).
Mohaghegh, P. & Hickson, I. D. DNA helicase deficiencies associated with cancer predisposition and premature ageing disorders. Hum. Mol. Genet. 10, 741–746 (2001).
Vorechovsky, I. et al. Clustering of missense mutations in the ataxia-telangiectasia gene in a sporadic T-cell leukaemia. Nature Genet. 17, 96–99 (1997).
Menisser-de Murcia, J., Mark, M., Wendling, O., Wynshaw-Boris, A. & de Murcia, G. Early embryonic lethality in PARP-1 Atm double-mutant mice suggests a functional synergy in cell proliferation during development. Mol. Cell Biol. 21, 1828–1832 (2001).
Bryant, H. E., Ying, S. & Helleday, T. Homologous recombination is involved in repair of chromium-induced DNA damage in mammalian cells. Mutat. Res. 599, 116–123 (2006).
Matsuura, S. et al. Positional cloning of the gene for Nijmegen breakage syndrome. Nature Genet. 19, 179–181 (1998).
Levine, A. J., Momand, J. & Finlay, C. A. The p53 tumour suppressor gene. Nature 351, 453–456 (1991).
Bell, D. W. et al. Heterozygous germ line hCHK2 mutations in Li–Fraumeni syndrome. Science 286, 2528–2531 (1999).
Cleaver, J. E. Cancer in xeroderma pigmentosum and related disorders of DNA repair. Nature Rev. Cancer 5, 564–573 (2005).
Cheng, L., Sturgis, E. M., Eicher, S. A., Spitz, M. R. & Wei, Q. Expression of nucleotide excision repair genes and the risk for squamous cell carcinoma of the head and neck. Cancer 94, 393–397 (2002).
Taniguchi, T. & D'Andrea, A. D. Molecular pathogenesis of Fanconi anemia: recent progress. Blood 107, 4223–4233 (2006).
Wang, L., Patel, U., Ghosh, L. & Banerjee, S. DNA polymerase β mutations in human colorectal cancer. Cancer Res. 52, 4824–4827 (1992).
Zheng, L. et al. Fen1 mutations result in autoimmunity, chronic inflammation and cancers. Nature Med. 13, 812–819 (2007).
Koster, D. A., Palle, K., Bot, E. S. M., Bjornsti, M.-A. and Dekker, N. H. Antitumour drugs impede DNA uncoiling by topoisomerase I. Nature 448, 213–217 (2007).
Wooster, R. et al. Localization of a breast cancer susceptibility gene, BRCA2, to chromosome 13q12-13. Science 265, 2088–2090 (1994).
Acknowledgements
We would like to thank the Medical Research Council, The Swedish Research Council, The Swedish Cancer Society, The Swedish Children's Cancer Foundation, The Swedish Pain Relief Foundation and Cancer Research UK for financial support. We recognize that we were unable to cover all aspects of DNA repair in cancer in this Review. We apologize to those whom we have been unable to cite owing to space constraints.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Patents regarding targeting DNA repair have been filed by the University of Sheffield's and University of Oxford's technology transfer companies with Thomas Helleday as named inventor.
Supplementary information
Supplementary information S1 (Table)
Synthetic lethal interactions of S. cerevisiae genes that are homologous to human DNA repair and checkpoint genes implicated in cancer. (PDF 710 kb)
Related links
Related links
DATABASES
ClinicalTrials.gov
National Cancer Institute
National Cancer Institute Drug Dictionary
FURTHER INFORMATION
Glossary
- Alkylating agents
-
Electrophilic compounds that are reactive either directly or following metabolism and bind covalently to electron-rich atoms in DNA bases (that is, oxygen and nitrogen).
- Antimetabolites
-
Compounds with similar chemical structures to nucleotide metabolites that interfere with nucleotide biosynthesis or are incorporated into DNA.
- Non-homologous end joining
-
Connection and resealing of the two ends of a DNA double-strand break without the need for sequence homology between the ends.
- Homologous recombination
-
A process that can copy a DNA sequence from an intact DNA molecule (often the newly synthesized sister chromatid) to repair or bypass replication lesions.
- Base-excision repair
-
A repair pathway that replaces missing or modified DNA bases, such as those produced by alkylating agents or in spontaneously degraded DNA, with the correct DNA base.
- Nucleotide-excision repair
-
A process that removes large DNA adducts or base modifications that distort the double helix and uses the opposite strand as template for repair.
- Alkyltransferases
-
A class of enzymes that directly reverse DNA base modifications that are induced by alkylating agents by transferring the alkyl group from the base onto the protein.
- DNA dioxygenases
-
A class of enzymes that directly reverse DNA base methylations through an oxidation mechanism. The human DNA dioxygenase ABH2 is thought to act at replication forks.
- Mismatch repair
-
A process that acts during DNA replication to correct base-pairing errors made by the DNA polymerases.
- Translesion synthesis
-
A mechanism during DNA replication in which the standard DNA polymerase is temporarily exchanged for a specialized polymerase that can synthesize DNA across base damage on the template strand.
- Fanconi anaemia repair pathway
-
Proteins of this pathway, including BRCA2, are mutated in the hereditary disorder Fanconi anaemia (FA), resulting in hypersensitivity to inter-strand crosslinks. Evidence suggests that the FA pathway promotes the repair of stalled replication forks, possibly by activating HR and facilitating ATR- and ATM-dependent checkpoint signalling.
- Endonuclease-mediated repair
-
A repair pathway that introduces a DNA single-strand break in a DNA structure to facilitate continuous repair.
- RecQ-mediated repair
-
A repair pathway that unwinds complex DNA structure to facilitate repair.
- Therapeutic index
-
The therapeutic index describes the ability of a treatment strategy to kill cancer cells in preference to cells in normal tissues.
- Synthetic lethality
-
A genetic phenomenon in which the combination of two otherwise non-lethal mutations results in an inviable cell. Synthetic lethal phenotypes are indicative of an interaction between the products of the two mutant genes within the cell.
- Biomarkers
-
A molecule or substance whose detection indicates a particular disease state or treatment response.
- Hypoxia
-
A subnormal concentration of oxygen. In cancer tissue, hypoxia is often the result of abnormal vasculature.
Rights and permissions
About this article
Cite this article
Helleday, T., Petermann, E., Lundin, C. et al. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer 8, 193–204 (2008). https://doi.org/10.1038/nrc2342
Issue Date:
DOI: https://doi.org/10.1038/nrc2342