Abstract
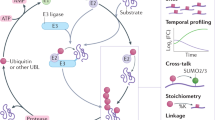
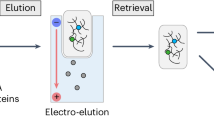
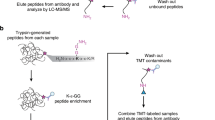
Ubiquitin and ubiquitin-like modifiers (UBLs) such as small ubiquitin-like modifier (SUMO) can act as antagonists to one another by competing to occupy similar residues in the proteome. In addition, SUMO and ubiquitin can be coupled to each other at key lysine residues to form highly branched protein networks. The interplay between these modifications governs important biological processes such as double-strand break repair and meiotic recombination. We recently developed an approach that permits the identification of proteins that are modified by both SUMOylation and ubiquitylation. This protocol requires cells that express a mutant 6×His-SUMO3m protein that has had its C terminus modified from QQQTGG to RNQTGG, enabling the purification of SUMOylated peptides and their identification by tandem mass spectrometry (MS/MS). Cells are lysed under denaturing conditions, and the SUMOylated proteins are purified on nickel-nitrilotriacetic acid (Ni-NTA) resin via the 6×His on the SUMO3m construct. After on-bead digestion using trypsin, ubiquitylated peptides are enriched by immunoprecipitation, and the flow-through from this step is subjected to anti-SUMO immunoprecipitation. The SUMOylated peptides are fractionated on strong cation exchange (SCX) StageTips to enhance the coverage of the SUMO proteome. The ubiquitylated and SUMOylated peptides are analyzed separately by liquid chromatography (LC)–MS/MS and identified with MaxQuant. We demonstrate how this approach can be used to identify temporal changes in SUMOylated and ubiquitylated proteins in response to, for instance, heat shock and proteasome inhibition. The procedure requires 3 d when starting from cell pellets and yields >8,000 SUMO sites and >3,500 ubiquitin sites from 16 mg of cell extract.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Bettermann, K., Benesch, M., Weis, S. & Haybaeck, J. SUMOylation in carcinogenesis. Cancer Lett. 316, 113–125 (2012).
Rodriguez, M.S., Dargemont, C. & Hay, R.T. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 276, 12654–12659 (2001).
Tatham, M.H. et al. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 276, 35368–35374 (2001).
Matic, I. et al. In vivo identification of human small ubiquitin-like modifier polymerization sites by high accuracy mass spectrometry and an in vitro to in vivo strategy. Mol. Cell. Proteomics 7, 132–144 (2008).
Tatham, M.H., Matic, I., Mann, M. & Hay, R.T. Comparative proteomic analysis identifies a role for SUMO in protein quality control. Sci. Signal. 4, rs4 (2011).
Geiss-Friedlander, R. & Melchior, F. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8, 947–956 (2007).
Bernier-Villamor, V., Sampson, D.A., Matunis, M.J. & Lima, C.D. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108, 345–356 (2002).
Creton, S. & Jentsch, S. SnapShot: the SUMO system. Cell 143, 848–848.e841 (2010).
Weisshaar, S.R. et al. Arsenic trioxide stimulates SUMO-2/3 modification leading to RNF4-dependent proteolytic targeting of PML. FEBS Lett. 582, 3174–3178 (2008).
Uzunova, K. et al. Ubiquitin-dependent proteolytic control of SUMO conjugates. J. Biol. Chem. 282, 34167–34175 (2007).
Geoffroy, M.-C. & Hay, R.T. An additional role for SUMO in ubiquitin-mediated proteolysis. Nat. Rev. Mol. Cell Biol. 10, 564–568 (2009).
Udeshi, N.D. et al. Methods for quantification of in vivo changes in protein ubiquitination following proteasome and deubiquitinase inhibition. Mol. Cell. Proteomics 11, 148–159 (2012).
Kim, W. et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 (2011).
Schwertman, P., Bekker-Jensen, S. & Mailand, N. Regulation of DNA double-strand break repair by ubiquitin and ubiquitin-like modifiers. Nat. Rev. Mol. Cell Biol. 17, 379–394 (2016).
Rao, H.B. et al. A SUMO-ubiquitin relay recruits proteasomes to chromosome axes to regulate meiotic recombination. Science 355, 403–407 (2017).
Hendriks, I.A. et al. Uncovering global SUMOylation signaling networks in a site-specific manner. Nat. Struct. Mol. Biol. 21, 927–936 (2014).
Lamoliatte, F. et al. Large-scale analysis of lysine SUMOylation by SUMO remnant immunoaffinity profiling. Nat. Commun. 5, 5409 (2014).
Tammsalu, T. et al. Proteome-wide identification of SUMO2 modification sites. Sci. Signal. 7, rs2 (2014).
Cox, J. et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 (2011).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008).
Galisson, F. et al. A novel proteomics approach to identify SUMOylated proteins and their modification sites in human cells. Mol. Cell. Proteomics 10, M110.004796 (2011).
McManus, F.P., Altamirano, C.D. & Thibault, P. In vitro assay to determine SUMOylation sites on protein substrates. Nat. Protoc. 11, 387–397 (2016).
Hendriks, I.A. & Vertegaal, A.C. A high-yield double-purification proteomics strategy for the identification of SUMO sites. Nat. Protoc. 11, 1630–1649 (2016).
Lamoliatte, F. et al. Targeted identification of SUMOylation sites in human proteins using affinity enrichment and paralog-specific reporter ions. Mol. Cell. Proteomics 12, 2536–2550 (2013).
Lamoliatte, F., McManus, F.P., Maarifi, G., Chelbi-Alix, M.K. & Thibault, P. Uncovering the SUMOylation and ubiquitylation crosstalk in human cells using sequential peptide immunopurification. Nat. Commun. 8, 14109 (2017).
Udeshi, N.D. et al. Refined preparation and use of anti-diglycine remnant (K-ɛ-GG) antibody enables routine quantification of 10,000s of ubiquitination sites in single proteomics experiments. Mol. Cell. Proteomics 12, 825–831 (2013).
Impens, F., Radoshevich, L., Cossart, P. & Ribet, D. Mapping of SUMO sites and analysis of SUMOylation changes induced by external stimuli. Proc. Natl Acad. Sci. USA 111, 12432–12437 (2014).
Tammsalu, T. et al. Proteome-wide identification of SUMO modification sites by mass spectrometry. Nat. Protoc. 10, 1374–1388 (2015).
Barysch, S.V., Dittner, C., Flotho, A., Becker, J. & Melchior, F. Identification and analysis of endogenous SUMO1 and SUMO2/3 targets in mammalian cells and tissues using monoclonal antibodies. Nat. Protoc. 9, 896–909 (2014).
Becker, J. et al. Detecting endogenous SUMO targets in mammalian cells and tissues. Nat. Struct. Mol. Biol. 20, 525–531 (2013).
Hendriks, I.A. & Vertegaal, A.C. A comprehensive compilation of SUMO proteomics. Nat. Rev. Mol. Cell Biol. 17, 581–595 (2016).
Batth, T.S., Francavilla, C. & Olsen, J.V. Off-line high-pH reversed-phase fractionation for in-depth phosphoproteomics. J. Proteome Res. 13, 6176–6186 (2014).
Svinkina, T. et al. Deep, quantitative coverage of the lysine acetylome using novel anti-acetyl-lysine antibodies and an optimized proteomic workflow. Mol. Cell. Proteomics 14, 2429–2440 (2015).
Udeshi, N.D., Mertins, P., Svinkina, T. & Carr, S.A. Large-scale identification of ubiquitination sites by mass spectrometry. Nat. Protoc. 8, 1950–1960 (2013).
Vizcaino, J.A. et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, 11033 (2016).
Rappsilber, J., Mann, M. & Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 (2007).
Acknowledgements
This work was funded by the Natural Sciences and Engineering Research Council of Canada (NSERC) and Genome Canada. F.P.M. is the recipient of a postdoctoral fellowship from NSERC. F.L. was supported by a scholarship from the faculty of graduate studies of the Université de Montreal. The Institute for Research in Immunology and Cancer (IRIC) receives infrastructure support from Genome Canada and Génome Québec, the Institute for Research in Immunology and Cancer — Commercialization of Research (IRICoR), the Canadian Foundation for Innovation, and the Fonds de Recherche du Québec – Santé (FRQS).
Author information
Authors and Affiliations
Contributions
F.P.M. and F.L. carried out the experiments. F.P.M., F.L. and P.T. wrote the manuscript. P.T. developed the concept and managed the project. F.P.M. and F.L. contributed equally to this work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
McManus, F., Lamoliatte, F. & Thibault, P. Identification of cross talk between SUMOylation and ubiquitylation using a sequential peptide immunopurification approach. Nat Protoc 12, 2354–2355 (2017). https://doi.org/10.1038/nprot.2017.105
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2017.105
This article is cited by
-
Rhes, a striatal enriched protein, regulates post-translational small-ubiquitin-like-modifier (SUMO) modification of nuclear proteins and alters gene expression
Cellular and Molecular Life Sciences (2024)
-
Proteomic strategies for characterizing ubiquitin-like modifications
Nature Reviews Methods Primers (2021)
-
Quantitative SUMO proteomics identifies PIAS1 substrates involved in cell migration and motility
Nature Communications (2020)
-
Gas-Phase Enrichment of Multiply Charged Peptide Ions by Differential Ion Mobility Extend the Comprehensiveness of SUMO Proteome Analyses
Journal of the American Society for Mass Spectrometry (2018)