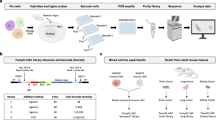
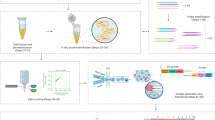
Abstract
We describe here a protocol for the representative amplification of global mRNAs from typical single mammalian cells to provide a template for high-density oligonucleotide microarray analysis. A single cell is lysed in a tube without purification and first-strand cDNAs are synthesized using a poly(dT)-tailed primer. Unreacted primer is specifically eliminated by exonuclease treatment and second strands are generated with a second poly(dT)-tailed primer after poly(dA) tailing of the first-strand cDNAs. The cDNAs are split into four tubes, which are independently directionally amplified by PCR, and then recombined. The amplified products (∼100 ng) show superior representation and reproducibility of original gene expression, especially for genes expressed in more than 20 copies per cell, compared with those obtained by a conventional PCR protocol, and can effectively be used for quantitative PCR and EST analyses. The cDNAs are then subjected to another PCR amplification with primers bearing the T7 promoter sequence. The resultant cDNA products are gel purified, amplified by one final cycle and used for isothermal linear amplification by T7 RNA polymerase to synthesize cRNAs for microarray hybridization. This protocol yields cDNA templates sufficient for more than 80 microarray hybridizations from a single cell, and can be completed in 5–6 days.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Lockhart, D.J. et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 14, 1675–1680 (1996).
Adams, M.D. et al. Complementary DNA sequencing: expressed sequence tags and human genome project. Science 252, 1651–1656 (1991).
Boguski, M.S., Lowe, T.M. & Tolstoshev, C.M. dbEST—database for “expressed sequence tags”. Nat. Genet. 4, 332–333 (1993).
Velculescu, V.E., Zhang, L., Vogelstein, B. & Kinzler, K.W. Serial analysis of gene expression. Science 270, 484–487 (1995).
Brady, G., Barbara, M. & Iscove, N. Representative in vitro cDNA amplification from individual hemopoietic cells and colonies. Methods Mol. Cell. Biol. 2, 17–25 (1990).
Brady, G. & Iscove, N.N. Construction of cDNA libraries from single cells. Methods Enzymol. 225, 611–623 (1993).
Van Gelder, R.N. et al. Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc. Natl. Acad. Sci. USA 87, 1663–1667 (1990).
Eberwine, J. et al. Analysis of gene expression in single live neurons. Proc. Natl. Acad. Sci. USA 89, 3010–3014 (1992).
Saito, H., Kubota, M., Roberts, R.W., Chi, Q. & Matsunami, H. RTP family members induce functional expression of mammalian odorant receptors. Cell 119, 679–691 (2004).
Klur, S., Toy, K., Williams, M.P. & Certa, U. Evaluation of procedures for amplification of small-size samples for hybridization on microarrays. Genomics 83, 508–517 (2004).
Ji, W., Zhou, W., Gregg, K., Lindpaintner, K. & Davis, S. A method for gene expression analysis by oligonucleotide arrays from minute biological materials. Anal. Biochem. 331, 329–339 (2004).
Iscove, N.N. et al. Representation is faithfully preserved in global cDNA amplified exponentially from sub-picogram quantities of mRNA. Nat. Biotechnol. 20, 940–943 (2002).
Kurimoto, K. et al. An improved single-cell cDNA amplification method for efficient high-density oligonucleotide microarray analysis. Nucleic Acids Res. 34, e42 (2006).
Tietjen, I. et al. Single-cell transcriptional analysis of neuronal progenitors. Neuron 38, 161–175 (2003).
Chazaud, C., Yamanaka, Y., Pawson, T. & Rossant, J. Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2–MAPK pathway. Dev. Cell 10, 615–624 (2006).
Baugh, L.R., Hill, A.A., Slonim, D.K., Brown, E.L. & Hunter, C.P. Composition and dynamics of the Caenorhabditis elegans early embryonic transcriptome. Development 130, 889–900 (2003).
Adams, M.D. et al. The genome sequence of Drosophila melanogaster . Science 287, 2185–2195 (2000).
Church, R.B. & Robertson, F.W. Biochemical analysis of genetic differences in the growth of Drosophila . Genet. Res. 7, 383–407 (1966).
Rahner, C., Fukuhara, M., Peng, S., Kojima, S. & Rizzolo, L.J. The apical and basal environments of the retinal pigment epithelium regulate the maturation of tight junctions during development. J. Cell Sci. 117, 3307–3318 (2004).
Jost, J.P., Ohno, T., Panyim, S. & Schuerch, A.R. Appearance of vitellogenin mRNA sequences and rate of vitellogenin synthesis in chicken liver following primary and secondary stimulation by 17 beta-estradiol. Eur. J. Biochem. 84, 355–361 (1978).
Brown, D.D. & Gurdon, J.B. Absence of ribosomal RNA aynthesis in the anucleolate mutant of Xenopus laevis . Proc. Natl. Acad. Sci. USA 51, 139–146 (1964).
Dulac, C. & Axel, R. A novel family of genes encoding putative pheromone receptors in mammals. Cell 83, 195–206 (1995).
Saitou, M., Barton, S.C. & Surani, M.A. A molecular programme for the specification of germ cell fate in mice. Nature 418, 293–300 (2002).
Chiang, M.K. & Melton, D.A. Single-cell transcript analysis of pancreas development. Dev. Cell 4, 383–393 (2003).
Yabuta, Y., Kurimoto, K., Ohinata, Y., Seki, Y. & Saitou, M. Gene expression dynamics during germline specification in mice identified by quantitative single-cell gene expression profiling. Biol. Reprod. 75, 705–716 (2006).
Kamme, F. et al. Single-cell microarray analysis in hippocampus CA1: demonstration and validation of cellular heterogeneity. J. Neurosci. 23, 3607–3615 (2003).
Seipp, S. & Buselmaier, W. Isolation of glyceraldehyde 3-phosphate dehydrogenase (Gapdh) cDNA from the distal half of mouse chromosome 16: further indication of a link between Alzheimer's disease and glycolysis. Neurosci. Lett. 182, 91–94 (1994).
Chambers, I. et al. Functional expression cloning of nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113, 643–655 (2003).
Mitsui, K. et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113, 631–642 (2003).
Avilion, A.A. et al. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 17, 126–140 (2003).
Takahashi, K., Mitsui, K. & Yamanaka, S. Role of ERas in promoting tumour-like properties in mouse embryonic stem cells. Nature 423, 541–545 (2003).
Nichols, J. et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95, 379–391 (1998).
Sato, M. et al. Identification of PGC7, a new gene expressed specifically in preimplantation embryos and germ cells. Mech. Dev. 113, 91–94 (2002).
Brennan, J. et al. Nodal signalling in the epiblast patterns the early mouse embryo. Nature 411, 965–969 (2001).
Rogers, M.B., Hosler, B.A. & Gudas, L.J. Specific expression of a retinoic acid-regulated, zinc-finger gene, Rex-1, in preimplantation embryos, trophoblast and spermatocytes. Development 113, 815–824 (1991).
Laible, G. et al. Mammalian homologues of the Polycomb-group gene Enhancer of zeste mediate gene silencing in Drosophila heterochromatin and at S. cerevisiae telomeres. EMBO J. 16, 3219–3232 (1997).
Kawakami, A. et al. Identification and functional characterization of a TIA-1-related nucleolysin. Proc. Natl. Acad. Sci. USA 89, 8681–8685 (1992).
Astigiano, S. et al. Changes in gene expression following exposure of nulli-SCCl murine embryonal carcinoma cells to inducers of differentiation: characterization of a down-regulated mRNA. Differentiation 46, 61–67 (1991).
MacGregor, G.R., Zambrowicz, B.P. & Soriano, P. Tissue non-specific alkaline phosphatase is expressed in both embryonic and extraembryonic lineages during mouse embryogenesis but is not required for migration of primordial germ cells. Development 121, 1487–1496 (1995).
Safrany, G. & Perry, R.P. Characterization of the mouse gene that encodes the delta/YY1/NF-E1/UCRBP transcription factor. Proc. Natl. Acad. Sci. USA 90, 5559–5563 (1993).
Morrisey, E.E., Ip, H.S., Lu, M.M. & Parmacek, M.S. GATA-6: a zinc finger transcription factor that is expressed in multiple cell lineages derived from lateral mesoderm. Dev. Biol. 177, 309–322 (1996).
Acknowledgements
We thank all the members of our laboratory for their discussion of this study. This study was supported in part by a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and by a PRESTO project grant from the Japan Science and Technology Agency.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Kurimoto, K., Yabuta, Y., Ohinata, Y. et al. Global single-cell cDNA amplification to provide a template for representative high-density oligonucleotide microarray analysis. Nat Protoc 2, 739–752 (2007). https://doi.org/10.1038/nprot.2007.79
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2007.79
This article is cited by
-
ARID1A promotes genomic stability through protecting telomere cohesion
Nature Communications (2019)
-
Cell-cycle-independent transitions in temporal identity of mammalian neural progenitor cells
Nature Communications (2016)
-
A developmental coordinate of pluripotency among mice, monkeys and humans
Nature (2016)
-
Laminin-guided highly efficient endothelial commitment from human pluripotent stem cells
Scientific Reports (2016)
-
Using single nuclei for RNA-seq to capture the transcriptome of postmortem neurons
Nature Protocols (2016)