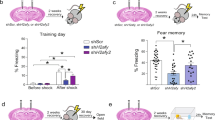
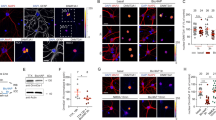
Abstract
The ability to form memories is a prerequisite for an organism's behavioral adaptation to environmental changes. At the molecular level, the acquisition and maintenance of memory requires changes in chromatin modifications. In an effort to unravel the epigenetic network underlying both short- and long-term memory, we examined chromatin modification changes in two distinct mouse brain regions, two cell types and three time points before and after contextual learning. We found that histone modifications predominantly changed during memory acquisition and correlated surprisingly little with changes in gene expression. Although long-lasting changes were almost exclusive to neurons, learning-related histone modification and DNA methylation changes also occurred in non-neuronal cell types, suggesting a functional role for non-neuronal cells in epigenetic learning. Finally, our data provide evidence for a molecular framework of memory acquisition and maintenance, wherein DNA methylation could alter the expression and splicing of genes involved in functional plasticity and synaptic wiring.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Guzman-Karlsson, M.C., Meadows, J.P., Gavin, C.F., Hablitz, J.J. & Sweatt, J.D. Transcriptional and epigenetic regulation of Hebbian and non-Hebbian plasticity. Neuropharmacology 80, 3–17 (2014).
Gräff, J. & Tsai, L.-H. Histone acetylation: molecular mnemonics on the chromatin. Nat. Rev. Neurosci. 14, 97–111 (2013).
Zovkic, I.B., Guzman-Karlsson, M.C. & Sweatt, J.D. Epigenetic regulation of memory formation and maintenance. Learn. Mem. 20, 61–74 (2013).
Sweatt, J.D. The emerging field of neuroepigenetics. Neuron 80, 624–632 (2013).
Lopez-Atalaya, J.P. & Barco, A. Can changes in histone acetylation contribute to memory formation? Trends Genet. 30, 529–539 (2014).
Levenson, J.M. et al. Regulation of histone acetylation during memory formation in the hippocampus. J. Biol. Chem. 279, 40545–40559 (2004).
Gupta, S. et al. Histone methylation regulates memory formation. J. Neurosci. 30, 3589–3599 (2010).
Miller, C.A. & Sweatt, J.D. Covalent modification of DNA regulates memory formation. Neuron 53, 857–869 (2007).
Miller, C.A. et al. Cortical DNA methylation maintains remote memory. Nat. Neurosci. 13, 664–666 (2010).
Strahl, B.D. & Allis, C.D. The language of covalent histone modifications. Nature 403, 41–45 (2000).
Day, J.J. & Sweatt, J.D. Epigenetic mechanisms in cognition. Neuron 70, 813–829 (2011).
Fischer, A. Epigenetic memory: the Lamarckian brain. EMBO J. 33, 945–967 (2014).
Fanselow, M.S. Factors governing one-trial contextual conditioning. Anim. Learn. Behav. 18, 264–270 (1990).
Kim, J.J. & Fanselow, M.S. Modality-specific retrograde amnesia of fear. Science 256, 675–677 (1992).
Runyan, J.D., Moore, A.N. & Dash, P.K. A role for prefrontal cortex in memory storage for trace fear conditioning. J. Neurosci. 24, 1288–1295 (2004).
Einarsson, E.O. & Nader, K. Involvement of the anterior cingulate cortex in formation, consolidation, and reconsolidation of recent and remote contextual fear memory. Learn. Mem. 19, 449–452 (2012).
Bonn, S. et al. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat. Genet. 44, 148–156 (2012).
Bonn, S. et al. Cell type-specific chromatin immunoprecipitation from multicellular complex samples using BiTS-ChIP. Nat. Protoc. 7, 978–994 (2012).
Jiang, Y., Matevossian, A., Huang, H.-S., Straubhaar, J. & Akbarian, S. Isolation of neuronal chromatin from brain tissue. BMC Neurosci. 9, 42 (2008).
Peleg, S. et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science 328, 753–756 (2010).
Barski, A. et al. High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 (2007).
Zhou, V.W., Goren, A. & Bernstein, B.E. Charting histone modifications and the functional organization of mammalian genomes. Nat. Rev. Genet. 12, 7–18 (2010).
Cahoy, J.D. et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci. 28, 264–278 (2008).
Ko, Y. et al. Cell type-specific genes show striking and distinct patterns of spatial expression in the mouse brain. Proc. Natl. Acad. Sci. USA 110, 3095–3100 (2013).
Guo, J.U. et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat. Neurosci. 14, 1345–1351 (2011).
Guo, J.U. et al. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat. Neurosci. 17, 215–222 (2014).
Lister, R. et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462, 315–322 (2009).
Kim, T.-K. et al. Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182–187 (2010).
Malik, A.N. et al. Genome-wide identification and characterization of functional neuronal activity-dependent enhancers. Nat. Neurosci. 17, 1330–1339 (2014).
Heintzman, N.D. et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459, 108–112 (2009).
Lyons, G.E., Micales, B.K., Schwarz, J., Martin, J.F. & Olson, E.N. Expression of mef2 genes in the mouse central nervous system suggests a role in neuronal maturation. J. Neurosci. 15, 5727–5738 (1995).
Gao, Z. et al. Neurod1 is essential for the survival and maturation of adult-born neurons. Nat. Neurosci. 12, 1090–1092 (2009).
Nakayama, A. et al. Role for RFX transcription factors in non-neuronal cell-specific inactivation of the microtubule-associated protein MAP1A promoter. J. Biol. Chem. 278, 233–240 (2003).
Reiprich, S. & Wegner, M. From CNS stem cells to neurons and glia: Sox for everyone. Cell Tissue Res. 359, 111–124 (2015).
Lesburguères, E. et al. Early tagging of cortical networks is required for the formation of enduring associative memory. Science 331, 924–928 (2011).
Reijmers, L.G., Perkins, B.L., Matsuo, N. & Mayford, M. Localization of a stable neural correlate of associative memory. Science 317, 1230–1233 (2007).
Garner, A.R. et al. Generation of a synthetic memory trace. Science 335, 1513–1516 (2012).
Silva, A.J., Zhou, Y., Rogerson, T., Shobe, J. & Balaji, J. Molecular and cellular approaches to memory allocation in neural circuits. Science 326, 391–395 (2009).
Ramirez, S. et al. Creating a false memory in the hippocampus. Science 341, 387–391 (2013).
Park, C.S., Rehrauer, H. & Mansuy, I.M. Genome-wide analysis of H4K5 acetylation associated with fear memory in mice. BMC Genomics 14, 539 (2013).
Bero, A.W. et al. Early remodeling of the neocortex upon episodic memory encoding. Proc. Natl. Acad. Sci. USA 111, 11852–11857 (2014).
Heyward, F.D. & Sweatt, J.D. DNA methylation in memory formation: emerging insights. Neuroscientist 21, 475–489 (2015).
Shukla, S. et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 479, 74–79 (2011).
Fosque, B.F. et al. Neural circuits. Labeling of active neural circuits in vivo with designed calcium integrators. Science 347, 755–760 (2015).
Peng, X. et al. Statistical implications of pooling RNA samples for microarray experiments. BMC Bioinformatics 4, 26 (2003).
Kendziorski, C., Irizarry, R.A., Chen, K.S., Haag, J.D. & Gould, M.N. On the utility of pooling biological samples in microarray experiments. Proc. Natl. Acad. Sci. USA 102, 4252–4257 (2005).
Kendziorski, C.M., Zhang, Y., Lan, H. & Attie, A.D. The efficiency of pooling mRNA in microarray experiments. Biostatistics 4, 465–477 (2003).
Egelhofer, T.A. et al. An assessment of histone-modification antibody quality. Nat. Struct. Mol. Biol. 18, 91–93 (2011).
Proudhon, C. et al. Protection against de novo methylation is instrumental in maintaining parent-of-origin methylation inherited from the gametes. Mol. Cell 47, 909–920 (2012).
Bessa, J. et al. Zebrafish enhancer detection (ZED) vector: a new tool to facilitate transgenesis and the functional analysis of cis-regulatory regions in zebrafish. Dev. Dyn. 238, 2409–2417 (2009).
Kimmel, C.B., Ballard, W.W., Kimmel, S.R., Ullmann, B. & Schilling, T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310 (1995).
Andrews, S. FastQC A quality control tool for high throughput sequence data http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (2010).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Lienhard, M., Grimm, C., Morkel, M., Herwig, R. & Chavez, L. MEDIPS: genome-wide differential coverage analysis of sequencing data derived from DNA enrichment experiments. Bioinformatics 30, 284–286 (2014).
Capece, V. et al. Oasis: online analysis of small RNA deep sequencing data. Bioinformatics 31, 2205–2207 (2015).
Landt, S.G. et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 22, 1813–1831 (2012).
Salzberg, S.L. & Langmead, B. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Shen, L., Shao, N., Liu, X. & Nestler, E. ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics 15, 284 (2014).
Love, M.I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Wang, J., Duncan, D., Shi, Z. & Zhang, B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 41, W77–W83 (2013).
Rajagopal, N. et al. RFECS: a random-forest based algorithm for enhancer identification from chromatin state. PLoS Comput. Biol. 9, e1002968 (2013).
Quinlan, A.R. & Hall, I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Song, Q. & Smith, A.D. Identifying dispersed epigenomic domains from ChIP-Seq data. Bioinformatics 27, 870–871 (2011).
Anders, S., Reyes, A. & Huber, W. Detecting differential usage of exons from RNA-seq data. Genome Res. 22, 2008–2017 (2012).
Acknowledgements
We would like to thank M. Boroomandi for help with the behavioral experiments. We would like to thank E.E. Furlong, A.G. Ladurner, C. Margulies, R.P. Zinzen and W. Jackson for critical reading of the manuscript. This work was supported by the DFG (BO4224/4-1) (S. Bonn), the Network of Centres of Excellence in Neurodegeneration (CoEN) initiative (S. Bonn and A.F.), iMed – the Helmholtz Initiative on Personalized Medicine (S. Bonn and A.F.), the EURYI Award of the ESF (A.F.), the Hans and Ilse Breuer Foundation (A.F.), and by the European Research Council under the European Union's Seventh Framework Program (FP7/2007–2013)/ ERC Grant Agreement No. 321366-Amyloid (advanced grant to C.H.).
Author information
Authors and Affiliations
Contributions
S. Bonn initiated the study and designed the experiments with A.F., R.H., M.H. and R.O.V. S. Burkhardt, M.N.S., R.H. and S.B.J. performed the behavioral experiments. F.v.B., C.H. and B.S. performed the zebrafish experiments. A.-L.S., S. Burkhardt, R.H. and M.H. were responsible for the library generation and sequencing of the samples. M.H., R.H., A.R. and E.B. conducted all other experiments and analyzed the data. R.O.V., O.S., R.-U.R., T.P.C., J.C.G.V., V.C., S. Bonn, R.H. and M.H. were responsible for the computational analysis of the data. S. Bonn, M.H. and R.O.V. wrote the manuscript. All of the authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–30 (PDF 9692 kb)
Supplementary Table 1: ChIP- and MeDIP-seq antibodies.
Detailed information on the antibodies used for ChIP- and MeDIP-seq experiments and their usage. (XLS 29 kb)
Supplementary Table 2: DEGs and DEEs.
Overview over all DEGs and DEEs for the different time-points, learning comparisons, and brain areas. Additional information on in vitro DEGs after KCl stimulation. (XLS 13418 kb)
Supplementary Table 3: Sequencing samples and quality.
Summarization of ChIP-, MeDIP-, and RNA-seq samples and their corresponding quality metrics. (XLS 88 kb)
Supplementary Table 4: Known cell type-specific genes.
Table containing the genes that were categorized as neuron- or glia-specific based on published information. (XLS 44 kb)
Supplementary Table 5: Predicted cell type-specific genes.
Table of the predicted cell-type specific genes in the CA1 and ACC, in neurons and non-neurons. (XLS 125 kb)
Supplementary Table 6: Predicted CRMs.
Table of the predicted cell-type specific CRMs in the CA1 and ACC, in neurons and non-neurons. (XLS 11301 kb)
Supplementary Table 7: Validated CRMs.
Detailed information on the CRMs that cloned and validated in Danio rerio enhancer assays. (XLS 53 kb)
Supplementary Table 8: Differential HPTMs.
Lists of genes containing DHPTMs for the in vivo and in vitro data and summary information. (XLS 158 kb)
Supplementary Table 9: Primer.
Table summarizing information of the ChIP, expression, and MeDIP qPCR primers used. (XLS 39 kb)
Supplementary Table 10: DHPTM-DEG overview.
Information on DHPTM-DEG comparisons for the different histone modifications, learning comparisons, and cell types. (XLS 57 kb)
Supplementary Table 11: DMRs and DMGs.
Overview over all DMRs for the different time-points, learning comparisons, brain areas, and cell types. (XLS 23253 kb)
Supplementary Table 12: DMG-DEG-DEE overview.
Information on DMG-DEG and DMG-DEE comparisons for the different time-points, learning comparisons, brain areas, and cell types. (XLS 430 kb)
Supplementary Data Set
Interactive Enrichment Files (ZIP 6838 kb)
Rights and permissions
About this article
Cite this article
Halder, R., Hennion, M., Vidal, R. et al. DNA methylation changes in plasticity genes accompany the formation and maintenance of memory. Nat Neurosci 19, 102–110 (2016). https://doi.org/10.1038/nn.4194
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.4194
This article is cited by
-
Formation of memory assemblies through the DNA-sensing TLR9 pathway
Nature (2024)
-
The Interplay Between Dietary Choline and Cardiometabolic Disorders: A Review of Current Evidence
Current Nutrition Reports (2024)
-
Hippocampal cells segregate positive and negative engrams
Communications Biology (2022)
-
Histone macroH2A1 is a stronger regulator of hippocampal transcription and memory than macroH2A2 in mice
Communications Biology (2022)
-
Altered m6A RNA methylation contributes to hippocampal memory deficits in Huntington’s disease mice
Cellular and Molecular Life Sciences (2022)