Abstract
The circadian (∼24 h) clock is continuously entrained (reset) by ambient light so that endogenous rhythms are synchronized with daily changes in the environment. Light-induced gene expression is thought to be the molecular mechanism underlying clock entrainment. mRNA translation is a key step of gene expression, but the manner in which clock entrainment is controlled at the level of mRNA translation is not well understood. We found that a light- and circadian clock–regulated MAPK/MNK pathway led to phosphorylation of the cap-binding protein eIF4E in the mouse suprachiasmatic nucleus of the hypothalamus, the locus of the master circadian clock in mammals. Phosphorylation of eIF4E specifically promoted translation of Period 1 (Per1) and Period 2 (Per2) mRNAs and increased the abundance of basal and inducible PER proteins, which facilitated circadian clock resetting and precise timekeeping. Together, these results highlight a critical role for light-regulated translational control in the physiology of the circadian clock.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Rosbash, M. The implications of multiple circadian clock origins. PLoS Biol. 7, e62 (2009).
Reppert, S.M. & Weaver, D.R. Coordination of circadian timing in mammals. Nature 418, 935–941 (2002).
Colwell, C.S. Linking neural activity and molecular oscillations in the SCN. Nat. Rev. Neurosci. 12, 553–569 (2011).
Takahashi, J.S., Hong, H.K., Ko, C.H. & McDearmon, E.L. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat. Rev. Genet. 9, 764–775 (2008).
Herzog, E.D. Neurons and networks in daily rhythms. Nat. Rev. Neurosci. 8, 790–802 (2007).
Peirson, S. & Foster, R.G. Melanopsin: another way of signaling light. Neuron 49, 331–339 (2006).
Golombek, D.A. & Rosenstein, R.E. Physiology of circadian entrainment. Physiol. Rev. 90, 1063–1102 (2010).
Albrecht, U., Sun, Z.S., Eichele, G. & Lee, C.C. A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell 91, 1055–1064 (1997).
Shearman, L.P., Zylka, M.J., Weaver, D.R., Kolakowski, L.F. Jr. & Reppert, S.M. Two period homologs: circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron 19, 1261–1269 (1997).
Shigeyoshi, Y. et al. Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript. Cell 91, 1043–1053 (1997).
Cao, R., Butcher, G.Q., Karelina, K., Arthur, J.S. & Obrietan, K. Mitogen- and stress-activated protein kinase 1 modulates photic entrainment of the suprachiasmatic circadian clock. Eur. J. Neurosci. 37, 130–140 (2013).
Travnickova-Bendova, Z., Cermakian, N., Reppert, S.M. & Sassone-Corsi, P. Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK/BMAL1 activity. Proc. Natl. Acad. Sci. USA 99, 7728–7733 (2002).
Sonenberg, N. & Hinnebusch, A.G. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136, 731–745 (2009).
Gingras, A.C., Raught, B. & Sonenberg, N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68, 913–963 (1999).
Cao, R. et al. Translational control of entrainment and synchrony of the suprachiasmatic circadian clock by mTOR/4E–BP1 signaling. Neuron 79, 712–724 (2013).
Buxade, M., Parra-Palau, J.L. & Proud, C.G. The Mnks: MAP kinase-interacting kinases (MAP kinase signal–integrating kinases). Front. Biosci. 13, 5359–5373 (2008).
Ueda, T., Watanabe-Fukunaga, R., Fukuyama, H., Nagata, S. & Fukunaga, R. Mnk2 and Mnk1 are essential for constitutive and inducible phosphorylation of eukaryotic initiation factor 4E but not for cell growth or development. Mol. Cell. Biol. 24, 6539–6549 (2004).
Furic, L. et al. eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proc. Natl. Acad. Sci. USA 107, 14134–14139 (2010).
Obrietan, K., Impey, S. & Storm, D.R. Light and circadian rhythmicity regulate MAP kinase activation in the suprachiasmatic nuclei. Nat. Neurosci. 1, 693–700 (1998).
Kelleher, R.J. III, Govindarajan, A., Jung, H.Y., Kang, H. & Tonegawa, S. Translational control by MAPK signaling in long-term synaptic plasticity and memory. Cell 116, 467–479 (2004).
Favata, M.F. et al. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J. Biol. Chem. 273, 18623–18632 (1998).
Knauf, U., Tschopp, C. & Gram, H. Negative regulation of protein translation by mitogen-activated protein kinase-interacting kinases 1 and 2. Mol. Cell. Biol. 21, 5500–5511 (2001).
Harbour, V.L., Robinson, B. & Amir, S. Variations in daily expression of the circadian clock protein, PER2, in the rat limbic forebrain during stable entrainment to a long light cycle. J. Mol. Neurosci. 45, 154–161 (2011).
Akiyama, M. et al. Inhibition of light- or glutamate-induced mPer1 expression represses the phase shifts into the mouse circadian locomotor and suprachiasmatic firing rhythms. J. Neurosci. 19, 1115–1121 (1999).
Albrecht, U., Zheng, B., Larkin, D., Sun, Z.S. & Lee, C.C. MPer1 and mper2 are essential for normal resetting of the circadian clock. J. Biol. Rhythms 16, 100–104 (2001).
Hastings, M.H., Field, M.D., Maywood, E.S., Weaver, D.R. & Reppert, S.M. Differential regulation of mPER1 and mTIM proteins in the mouse suprachiasmatic nuclei: new insights into a core clock mechanism. J. Neurosci. 19, RC11 (1999).
Yan, L. & Silver, R. Resetting the brain clock: time course and localization of mPER1 and mPER2 protein expression in suprachiasmatic nuclei during phase shifts. Eur. J. Neurosci. 19, 1105–1109 (2004).
Cao, R., Li, A., Cho, H.Y., Lee, B. & Obrietan, K. Mammalian target of rapamycin signaling modulates photic entrainment of the suprachiasmatic circadian clock. J. Neurosci. 30, 6302–6314 (2010).
Balsalobre, A., Damiola, F. & Schibler, U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93, 929–937 (1998).
Lee, C., Etchegaray, J.P., Cagampang, F.R., Loudon, A.S. & Reppert, S.M. Posttranslational mechanisms regulate the mammalian circadian clock. Cell 107, 855–867 (2001).
Konicek, B.W. et al. Therapeutic inhibition of MAP kinase interacting kinase blocks eukaryotic initiation factor 4E phosphorylation and suppresses outgrowth of experimental lung metastases. Cancer Res. 71, 1849–1857 (2011).
Tischkau, S.A.M.J., Tyan, S.H., Buchanan, G.F. & Gillette, M.U. Ca2+/cAMP response element-binding protein (CREB)-dependent activation of Per1 is required for light-induced signaling in the suprachiasmatic nucleus circadian clock. J. Biol. Chem. 278, 718–723 (2003).
Gamble, K.L., Allen, G.C., Zhou, T. & McMahon, D.G. Gastrin-releasing peptide mediates light-like resetting of the suprachiasmatic nucleus circadian pacemaker through cAMP response element-binding protein and Per1 activation. J. Neurosci. 27, 12078–12087 (2007).
Ginty, D.D. et al. Regulation of CREB phosphorylation in the suprachiasmatic nucleus by light and a circadian clock. Science 260, 238–241 (1993).
Jagannath, A. et al. The CRTC1–SIK1 pathway regulates entrainment of the circadian clock. Cell 154, 1100–1111 (2013).
Sakamoto, K. et al. Clock and light regulation of the CREB coactivator CRTC1 in the suprachiasmatic circadian clock. J. Neurosci. 33, 9021–9027 (2013).
Cheng, H.Y. et al. microRNA modulation of circadian-clock period and entrainment. Neuron 54, 813–829 (2007).
Alvarez-Saavedra, M. et al. miRNA-132 orchestrates chromatin remodeling and translational control of the circadian clock. Hum. Mol. Genet. 20, 731–751 (2011).
Fustin, J.M. et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 155, 793–806 (2013).
Meng, Q.J. et al. Setting clock speed in mammals: the CK1 epsilon tau mutation in mice accelerates circadian pacemakers by selectively destabilizing PERIOD proteins. Neuron 58, 78–88 (2008).
Kojima, S. et al. LARK activates posttranscriptional expression of an essential mammalian clock protein, PERIOD1. Proc. Natl. Acad. Sci. USA 104, 1859–1864 (2007).
Morf, J. et al. Cold-inducible RNA-binding protein modulates circadian gene expression posttranscriptionally. Science 338, 379–383 (2012).
Bradley, S., Narayanan, S. & Rosbash, M. NAT1/DAP5/p97 and atypical translational control in the Drosophila circadian oscillator. Genetics 192, 943–957 (2012).
Lim, C. et al. The novel gene twenty-four defines a critical translational step in the Drosophila clock. Nature 470, 399–403 (2011).
Lim, C. & Allada, R. ATAXIN-2 activates PERIOD translation to sustain circadian rhythms in Drosophila. Science 340, 875–879 (2013).
Zhang, Y., Ling, J., Yuan, C., Dubruille, R. & Emery, P. A role for Drosophila ATX2 in activation of PER translation and circadian behavior. Science 340, 879–882 (2013).
Thaben, P.F. & Westermark, P.O. Detecting rhythms in time series with RAIN. J. Biol. Rhythms 29, 391–400 (2014).
Acknowledgements
We thank D. Weaver, I. Edery and J. Stewart for advice and critical reading of the manuscript, and C. Zakaria, N. Robichaud, A. Sylvestre, B. Robinson and I. Harvey for excellent technical assistance. We are indebted to D. Weaver (University of Massachusetts Medical School) for his gift of the anti-PER1 serum and J. Takahashi (University of Texas Southwestern Medical Center) for his gift of the mPER2∷LUC transgenic mice. This work was supported by Canadian Institute of Health Research grants (MOP 114994 to N.S. and MOP 13625 to S.A.) and a US National Institutes of Health grant (NINDS R01NS054794 to A.C.L.). N.S. receives funding from the Howard Hughes Medical Institute. R.C. and N.d.Z. are recipients of the Fonds de recherche du Québec-Santé (FRQS) Postdoctoral Award and R.C. is a recipient of the Banting Postdoctoral Fellowship.
Author information
Authors and Affiliations
Contributions
R.C., S.A. and N.S. designed the study. R.C., N.d.Z., C.G.G., I.D.B., Y.T., A.Y. and H.X. performed the experiments. C.L. and K.-F.S. contributed reagents and analytic tools. R.C., I.D.B., C.G.G., A.Y. and H.X. analyzed the data. R.C., A.C.L., S.A. and N.S. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Light-induced phosphorylation of MNK1 in the SCN
(a) Representative microscopic images of coronal SCN sections immunolabeled for p-MNK1 (at Thr250) are shown. For these experiments, animals were entrained under a 12h/12h LD cycle for 14 d and transferred to DD for 2d. On the 3rd day in DD, mice were exposed to light (55 lx, 15 min) at CT15. Mice were sacrificed 30 min after light. Scale bars: 100 μm. (b) Quantitation of intensities of p-MNK1 labeling. Note that light triggered a modest but significant increase of p-MNK1 expression at CT 15. *P<0.05 vs. No light. (c) Quantitation of the levels of ERK, MNK1 and eIF4E in the SCN over a 24-h period (See Figure 1f for Western blot images). The protein levels are normalized to the level of α-tubulin. SCN-enriched tissue was harvested every 4 h throughout 24 h when mice were in DD. (d) Phosphorylation of ERK, eIF4E and total MNK1/2 in the SCN over a 24-h period. SCN-enriched tissue was harvested every 4 h throughout 24 h when mice were in LD. The blots of total ERK and eIF4E were used as control. Full-length blots/gels are presented in Supplementary Figure 6.
Supplementary Figure 2 Light-induced ERK phosphorylation and c-Fos expression in the SCN
(a) and (c) Representative micrographs of SCN tissue labeled for the phosphorylated (at Thr 202 and/or Tyr 204) form of ERK (p-ERK, a) and c-Fos (c). Relative to the control condition (No Light), photic stimulation (55 lux, 15 min) triggered a robust increase in ERK phosphorylation and c-Fos expression in the SCN of WT mice. Compared to the WT mice, light induced similar level of p-ERK and c-Fos expression in KI mice. For these experiments, mice were entrained to a 12 h/12 h LD cycle for 14 d and put in DD for 2 d. A light pulse (55 lux, 15 min) was applied at CT15. Animals were sacrificed 30 min after light. Scale bars: 100 μm. (b) and (d): Quantitation of light-induced ERK phosphorylation (b) and c-Fos expression (d). Light evoked significant p-ERK and c-Fos expression in the SCN of both WT and KI mice (*P<0.05, WT L vs. WT NL, KI L vs. KI NL by ANOVA). The p-ERK and c-Fos induction in the KI mice was not significantly different with that in the WT mice (P>0.05, KI L vs. WT L, by ANOVA). L: light; NL: no light
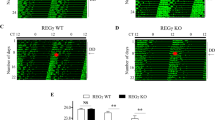
Supplementary Figure 3 Photic entrainment to non-24-h light/dark cycles (T-cycles) is impaired in the eIF4E KI mice
Representative actograms of wheel-running behavior demonstrating differential entrainment to a T21 (top), T26 (middle) and T27 (bottom) cycles in the WT and KI mice. Initially animals were kept in LD or DD for at least 7 d. A T-cycle was started and lasted for 14 d. Note that WT mice were more stably entrained by 14 d in the T21 or T27 cycles compared to the KI mice. However, both WT and KI mice were entrained to the T26 cycle.
Supplementary Figure 4 Inhibition of eIF4E phosphorylation decreases amplitudes of PER oscillations
(a) and (b) Temporal profiles of PER1(a) and PER2(b) in the SCN over 24 h. Representative microscopic images of SCN sections immunostained for PER1 (a) or PER2 (b) are shown. Quantitation of the staining intensity is shown in Figure 4a,b. For these experiments, animals were entrained for 14 d and dark-adapted for 2 d. On the 3rd day in DD, mice were sacrificed and brains were harvested every 4 h throughout 24 h. SCN sections were stained for PER1 or PER2. Note that the levels of PER1/2 proteins in the SCN exhibited circadian oscillations and reached a peak at around CT14 in both the WT and KI mice. However, PER1/2 levels at CT14 were decreased in the SCN of KI mice as compared to the WT mice. Scale bars: 100 μm. (c) Representative plots of mPER2::Luc bioluminescence patterns of SCN explants. Each plot was from one SCN explant. The period of the plots are quantified in (e). *P<0.05 (d) Representative plots of mPER2::Luc bioluminescence patterns of SCN explants before and after application of the MNK1 inhibitor CGP57380 (10 µM). Note that CGP57380 significantly decreased the amplitude of the mPER2::Luc rhythms in the WT but not in the KI SCN explant. Quantitation of the period is shown in (e) and the amplitude ratios (with CGP/without CGP) are shown in (f). Nine to thirteen animals were used for each group. *P<0.05 vs.WT
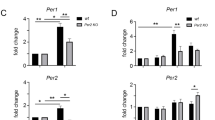
Supplementary Figure 5 Specific regulation of Period 1 and Period 2 mRNA translation by eIF4E phosphorylation
(a) and (b) Puromycin incorporation assay to measure global protein synthesis. For this experiment, animals were injected with puromycin (20 mg/kg, i.p.) and sacrificed 45 min after injection. Brains were harvested and processed for immunolabeling. (a) Representative images of puromycin labeling. Scale bar: 100 μm. (b) Quantitation of the labeling intensity reveals no significant difference in global protein synthesis rates between WT and KI SCN. (c) and (d) Effects of cercosporamide on Per expression. (c) Western blots indicating that the MNK inhibitor cercosporamide did not reduce PER protein levels in KI MEFs. Full-length blots/gels are presented in Supplementary Figure 6. (d) Cercosporamide does not inhibit translation of Per1 and Per2 5’UTR mRNA reporters. (e) Polysome profiles from forebrain lysates. From top to bottom: on the left are qRT-PCR results of levels of Clock, Bmal1, Cry1 and Cry2 mRNA extracted from the polysome fractions. qRT-PCR results on total mRNA extracted from the brain lysates are shown to the right. Note that no significant shift was detected in distribution profiles of Clock, Bmal1, Cry1 and Cry2 mRNAs. No difference was found in Clock, Bmal1, Cry1 and Cry2 mRNA levels between WT and KI mice (n=3, P>0.05 by Student’s t-test).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 and Supplementary Tables 1 and 2 (PDF 4415 kb)
Rights and permissions
About this article
Cite this article
Cao, R., Gkogkas, C., de Zavalia, N. et al. Light-regulated translational control of circadian behavior by eIF4E phosphorylation. Nat Neurosci 18, 855–862 (2015). https://doi.org/10.1038/nn.4010
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.4010