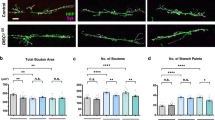
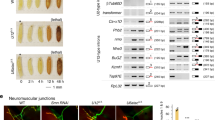
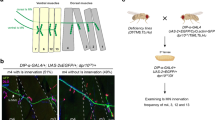
Abstract
Loss of FMR1 gene function results in fragile X syndrome, the most common heritable form of intellectual disability. The protein encoded by this locus (FMRP) is an RNA-binding protein that is thought to primarily act as a translational regulator; however, recent studies have implicated FMRP in other mechanisms of gene regulation. We found that the Drosophila fragile X homolog (dFMR1) biochemically interacted with the adenosine-to-inosine RNA-editing enzyme dADAR. Adar and Fmr1 mutant larvae exhibited distinct morphological neuromuscular junction (NMJ) defects. Epistasis experiments based on these phenotypic differences revealed that Adar acts downstream of Fmr1 and that dFMR1 modulates dADAR activity. Furthermore, sequence analyses revealed that a loss or overexpression of dFMR1 affects editing efficiency on certain dADAR targets with defined roles in synaptic transmission. These results link dFMR1 with the RNA-editing pathway and suggest that proper NMJ synaptic architecture requires modulation of dADAR activity by dFMR1.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
O'Donnell, W.T. & Warren, S.T. A decade of molecular studies of fragile X syndrome. Annu. Rev. Neurosci. 25, 315–338 (2002).
Feng, Y. et al. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol. Cell 1, 109–118 (1997).
Corbin, F. et al. The fragile X mental retardation protein is associated with poly(A)+ mRNA in actively translating polyribosomes. Hum. Mol. Genet. 6, 1465–1472 (1997).
Khandjian, E.W., Corbin, F., Woerly, S. & Rousseau, F. The fragile X mental retardation protein is associated with ribosomes. Nat. Genet. 12, 91–93 (1996).
Eberhart, D.E., Malter, H.E., Feng, Y. & Warren, S.T. The fragile X mental retardation protein is a ribonucleoprotein containing both nuclear localization and nuclear export signals. Hum. Mol. Genet. 5, 1083–1091 (1996).
Laggerbauer, B., Ostareck, D., Keidel, E.M., Ostareck-Lederer, A. & Fischer, U. Evidence that fragile X mental retardation protein is a negative regulator of translation. Hum. Mol. Genet. 10, 329–338 (2001).
Li, Z. et al. The fragile X mental retardation protein inhibits translation via interacting with mRNA. Nucleic Acids Res. 29, 2276–2283 (2001).
Monzo, K. et al. Fragile X mental retardation protein controls trailer hitch expression and cleavage furrow formation in Drosophila embryos. Proc. Natl. Acad. Sci. USA 103, 18160–18165 (2006).
Bechara, E.G. et al. A novel function for fragile X mental retardation protein in translational activation. PLoS Biol. 7, e16 (2009).
Weiler, I.J. et al. Fragile X mental retardation protein is translated near synapses in response to neurotransmitter activation. Proc. Natl. Acad. Sci. USA 94, 5395–5400 (1997).
Zalfa, F. et al. A new function for the fragile X mental retardation protein in regulation of PSD-95 mRNA stability. Nat. Neurosci. 10, 578–587 (2007).
Dictenberg, J.B., Swanger, S.A., Antar, L.N., Singer, R.H. & Bassell, G.J. A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. Dev. Cell 14, 926–939 (2008).
Estes, P.S., O'Shea, M., Clasen, S. & Zarnescu, D.C. Fragile X protein controls the efficacy of mRNA transport in Drosophila neurons. Mol. Cell. Neurosci. 39, 170–179 (2008).
Antar, L.N., Dictenberg, J.B., Plociniak, M., Afroz, R. & Bassell, G.J. Localization of FMRP-associated mRNA granules and requirement of microtubules for activity-dependent trafficking in hippocampal neurons. Genes Brain Behav. 4, 350–359 (2005).
Pepper, A.S., Beerman, R.W., Bhogal, B. & Jongens, T.A. Argonaute2 suppresses Drosophila fragile X expression preventing neurogenesis and oogenesis defects. PLoS ONE 4, e7618 (2009).
Jin, P. et al. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat. Neurosci. 7, 113–117 (2004).
Ishizuka, A., Siomi, M.C. & Siomi, H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 16, 2497–2508 (2002).
Caudy, A.A., Myers, M., Hannon, G.J. & Hammond, S.M. Fragile X–related protein and VIG associate with the RNA interference machinery. Genes Dev. 16, 2491–2496 (2002).
Xu, X.L., Li, Y., Wang, F. & Gao, F.B. The steady-state level of the nervous system–specific microRNA-124a is regulated by dFMR1 in Drosophila. J. Neurosci. 28, 11883–11889 (2008).
Edbauer, D. et al. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron 65, 373–384 (2010).
Bass, B.L. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 71, 817–846 (2002).
Palladino, M.J., Keegan, L.P., O'Connell, M.A. & Reenan, R.A. A-to-I pre-mRNA editing in Drosophila is primarily involved in adult nervous system function and integrity. Cell 102, 437–449 (2000).
Tonkin, L.A. et al. RNA editing by ADARs is important for normal behavior in Caenorhabditis elegans. EMBO J. 21, 6025–6035 (2002).
Higuchi, M. et al. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 406, 78–81 (2000).
Nishikura, K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 79, 321–349 (2010).
Zhang, Y.Q. et al. Drosophila fragile X–related gene regulates the MAP1B homolog Futsch to control synaptic structure and function. Cell 107, 591–603 (2001).
Veraksa, A., Bauer, A. & Artavanis-Tsakonas, S. Analyzing protein complexes in Drosophila with tandem affinity purification–mass spectrometry. Dev. Dyn. 232, 827–834 (2005).
Tsai, A. & Carstens, R.P. An optimized protocol for protein purification in cultured mammalian cells using a tandem affinity purification approach. Nat. Protoc. 1, 2820–2827 (2006).
Palladino, M.J., Keegan, L.P., O'Connell, M.A. & Reenan, R.A. dADAR, a Drosophila double-stranded RNA-specific adenosine deaminase is highly developmentally regulated and is itself a target for RNA editing. RNA 6, 1004–1018 (2000).
Jepson, J.E. et al. Engineered alterations in RNA editing modulate complex behavior in Drosophila: regulatory diversity of adenosine deaminase acting on RNA (ADAR) targets. J. Biol. Chem. 286, 8325–8337 (2011).
Devys, D., Lutz, Y., Rouyer, N., Bellocq, J.P. & Mandel, J.L. The FMR-1 protein is cytoplasmic, most abundant in neurons and appears normal in carriers of a fragile X premutation. Nat. Genet. 4, 335–340 (1993).
Feng, Y. et al. Fragile X mental retardation protein: nucleocytoplasmic shuttling and association with somatodendritic ribosomes. J. Neurosci. 17, 1539–1547 (1997).
Tamanini, F. et al. Differential expression of FMR1, FXR1 and FXR2 proteins in human brain and testis. Hum. Mol. Genet. 6, 1315–1322 (1997).
Hoopengardner, B., Bhalla, T., Staber, C. & Reenan, R. Nervous system targets of RNA editing identified by comparative genomics. Science 301, 832–836 (2003).
Koh, Y.H., Gramates, L.S. & Budnik, V. Drosophila larval neuromuscular junction: molecular components and mechanisms underlying synaptic plasticity. Microsc. Res. Tech. 49, 14–25 (2000).
Gatto, C.L. & Broadie, K. Temporal requirements of the fragile X mental retardation protein in the regulation of synaptic structure. Development 135, 2637–2648 (2008).
Keegan, L.P. et al. Tuning of RNA editing by ADAR is required in Drosophila. EMBO J. 24, 2183–2193 (2005).
Gallo, A., Keegan, L.P., Ring, G.M. & O'Connell, M.A. An ADAR that edits transcripts encoding ion channel subunits functions as a dimer. EMBO J. 22, 3421–3430 (2003).
Banerjee, P. et al. Substitution of critical isoleucines in the KH domains of Drosophila fragile X protein results in partial loss-of-function phenotypes. Genetics 175, 1241–1250 (2007).
Darnell, J.C. et al. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell 107, 489–499 (2001).
Darnell, J.C. et al. Kissing complex RNAs mediate interaction between the Fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes Dev. 19, 903–918 (2005).
Zang, J.B. et al. A mouse model of the human Fragile X syndrome I304N mutation. PLoS Genet. 5, e1000758 (2009).
Wan, L., Dockendorff, T.C., Jongens, T.A. & Dreyfuss, G. Characterization of dFMR1, a Drosophila melanogaster homolog of the fragile X mental retardation protein. Mol. Cell. Biol. 20, 8536–8547 (2000).
Dickman, D.K., Lu, Z., Meinertzhagen, I.A. & Schwarz, T.L. Altered synaptic development and active zone spacing in endocytosis mutants. Curr. Biol. 16, 591–598 (2006).
Aravamudan, B., Fergestad, T., Davis, W.S., Rodesch, C.K. & Broadie, K. Drosophila UNC-13 is essential for synaptic transmission. Nat. Neurosci. 2, 965–971 (1999).
Zhang, B. et al. Synaptic vesicle size and number are regulated by a clathrin adaptor protein required for endocytosis. Neuron 21, 1465–1475 (1998).
Dockendorff, T.C. et al. Drosophila lacking dfmr1 activity show defects in circadian output and fail to maintain courtship interest. Neuron 34, 973–984 (2002).
Puig, O. et al. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24, 218–229 (2001).
Rigaut, G. et al. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17, 1030–1032 (1999).
Zeng, F. et al. A protocol for PAIR: PNA-assisted identification of RNA binding proteins in living cells. Nat. Protoc. 1, 920–927 (2006).
Acknowledgements
We thank current and former members of the Jongens laboratory for helpful discussions and technical support, especially R. Beerman for preliminary studies with the LMB experiments, and S. Cruz and C. Dubowy. We also thank M. Sundaram and D. Hancks for critical reading of the manuscript. We are grateful to R. Carstens, G. Bashaw, T. Dockendorff, C. Hughes, K. Kaestner, S. Artavanis-Tsakonas, the Bloomington Stock Center and the Developmental Studies Hybridoma Bank for fly strains and reagents. We also thank C.-X. Yuan and The Perelman School of Medicine at the University of Pennsylvania Proteomics Facility for help with the mass spectrometry results. This work was supported by a Predoctoral Training grant in Genetics (5T32GM008216 to B.B.), a National Institute of Mental Health grant (MH086705 to T.A.J.), a National Institute of General Medical Science grant (GM086902 to T.A.J.) and an Ellison Medical Foundation Senior Scholar award to R.A.R.
Author information
Authors and Affiliations
Contributions
B.B. performed the experiments shown in Figures 1,2,3,4,5,6 and 7 and Supplementary Figures 2–5, generated the figures and wrote the manuscript. J.E.J. and Y.A.S. generated the Adar-HA4.5.2 and Adar-HA12.5.2 fly lines used in this study, performed experiments shown in Figure 7, helped generate the figure and contributed to the writing of this manuscript. J.E.J. also performed experiments for, and generated part of, Supplementary Figure 4. A.S.-R.P. performed the experiments shown in Supplementary Figure 1, generated the figure, identified the initial findings for this manuscript and contributed to the writing of this manuscript. R.A.R. and T.A.J. contributed to the conclusions drawn from all figures, provided intellectual input, contributed portions of the funding and contributed to the writing of this manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 and Supplementary Table 1 (PDF 9572 kb)
Rights and permissions
About this article
Cite this article
Bhogal, B., Jepson, J., Savva, Y. et al. Modulation of dADAR-dependent RNA editing by the Drosophila fragile X mental retardation protein. Nat Neurosci 14, 1517–1524 (2011). https://doi.org/10.1038/nn.2950
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.2950